Abstract
Background and Objectives
Resistance to the antiplatelet treatment with clopidogrel has both genetic and non-genetic causes. Polymorphic variants of cytochrome P450 3A4 isoenzyme involved in the bioactivation of clopidogrel might have an influence on responsiveness to the drug. The aim of this study was to evaluate the influence of CYP3A4*1G (IVS10+12G>A, rs2242480) on the pharmacokinetics and pharmacodynamics of clopidogrel.
Methods
CYP3A4*1G polymorphism was determined in a group of 82 patients undergoing percutaneous coronary intervention and taking 75 mg of clopidogrel daily. Concentrations of clopidogrel and its metabolites, inactive carboxylic acid derivative and two diastereoisomers of active thiol metabolite: H3 and H4, were determined by a validated HPLC–MS/MS method. Pharmacodynamic effect was measured by an impedance method with a Multiplate analyzer. Moreover, an effect of factors, such as CYP2C19 phenotype, age, gender, body mass index and interactions with drugs metabolized by CYP3A4 were also investigated.
Results
In the studied group allele frequencies were: wt—0.921, *1G—0.079. Pharmacokinetic parameters of clopidogrel and its metabolites were not significantly different in carriers of *1G allele, comparing to wt/wt homozygotes. Platelet aggregation was higher in heterozygotes than in wt/wt carriers; however, the difference was not statistically significant (p = 0.484). In a multivariate analysis, which included age, body mass index, co-morbidities and coadministered drugs, CYP3A4*1G was not a predictor of values of H3 and H4 pharmacokinetic parameters and platelet aggregation.
Conclusion
CYP3A4*1G might not be a significant contributor to the variability in pharmacokinetic and pharmacodynamic response to clopidogrel therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
No statistically significant differences in blood aggregation and pharmacokinetics of clopidogrel and its metabolites were found between CYP3A4*1G carriers and non-carriers. |
1 Introduction
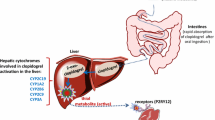
Clopidogrel (CLP) is a second-generation thienopyridine antiplatelet drug [1] Double antiplatelet therapy, which combines 75 mg of CLP and 75-325 mg of acetylsalicylic acid (aspirin) administered daily, is considered “a gold standard” in prevention of stent thrombosis after percutaneous coronary intervention [2]. However, 5–50 % of patients treated with CLP respond poorly to the drug [3, 4]. This phenomenon called “CLP resistance” is a main reason of the antiplatelet therapy failure, which may lead to ischaemic events and patient’s death. Many genetic and non-genetic factors are considered to be significant contributors to CLP resistance [5]. Among non-genetic factors are diabetes mellitus, impaired glucose tolerance, chronic kidney disease, ongoing inflammatory states, obesity and smoking. However, most widely discussed potential causes of CLP resistance are genetic polymorphisms of enzymes contributing to metabolic activation of CLP. CLP is a prodrug that is converted to the active, thiol metabolite in a two-step reaction catalyzed by CYP450 isoenzymes (CYP2C19, CYP3A4, CYP2B6, CYP1A2, CYP2C9) [6]. Thiol metabolite is detected in plasma as two diastereoisomers, H3 and H4, but only H4 exhibits antiaggregation properties [7]. Approximately 15 % of absorbed drug undergoes this metabolic pathway, while 85 % is hydrolyzed into an inactive carboxylic acid derivative (CLPM). Among CYP450 that take part in CLP bioactivation, CYP2C19 is of greatest importance. Many studies and meta-analyses have shown that carriers of CYP2C19 loss-of-function alleles are more susceptible to CLP resistance and high-on-treatment platelet reactivity and, therefore, to the occurrence of adverse cardiovascular events [8, 9]. According to Kazui et al. [6] CYP3A4 is responsible for transformation of 39.8 % of an intermediate product, 2-oxo-CLP, into the thiol entities. Therefore, altered activity of CYP3A4 resulting from genetic polymorphisms might have an influence on the yield of active metabolite formation and subsequently on the antiplatelet effect of the drug. Moreover, patients treated with CLP are often simultaneously treated with other drugs, such as statins or calcium channel blockers (CCB), that might be substrates of CYP3A4 and enhance or inhibit the function of this enzyme. Such interactions could have an influence on concentrations of H3 and H4 metabolites and the effectiveness on CLP treatment. Therefore, coadministration of those drugs cannot be neglected. In present study, we intended to evaluate influence of CYP3A4*1G (IVS10+12G>A, rs2242480) on CLP pharmacokinetics and pharmacodynamics. CYP3A4*1G is a single nucleotide polymorphism present in intron 10 of CYP3A4 with a possible enhancer and promoter activity [10]. Several studies have shown that the presence of CYP3A4*1G can be associated with altered pharmacokinetics of several drugs metabolized by CYP3A, such as fentanyl [11], atorvastatin [12] and tacrolimus [13]. So far only the influence of CYP3A4*1G on pharmacodynamic effect of CLP was evaluated [14–16]. To our knowledge this is the first study that investigates influence of CYP3A4*1G on pharmacokinetics of CLP in patients, with an emphasis on the active metabolite of CLP.
2 Materials and Methods
2.1 Study Group
A total number of 82 patients were recruited between 2011 and 2013 for participation in this observational study. All patients were of Caucasian origin and were scheduled for percutaneous coronary intervention (PCI) or coronarography. Patients were treated with 75 mg CLP daily for 7 days prior to the procedure to ensure achieving maximum level of platelet inhibition [17]. CLP was administered under fasting conditions. Exclusion criteria were acute myocardial infarction, malignancies, oral anticoagulation therapy with a coumarin derivative, treatment with a glycoprotein IIb/IIIa antagonist or other antiplatelet drugs except for aspirin, platelet count <100,000/μL, current liver dysfunction or impaired renal function (serum creatinine concentration 2 mg/dL).
2.2 Sample Collection
The sampling was performed on the seventh day of treatment with 75 mg of CLP. In 44 patients a pharmacokinetic profile of CLP and its metabolites was determined from samples collected 0.5, 1, 2, 3, 4, 6, 12 and 24 h after CLP administration. In 38 patients only 2 samples (0.5 and 2 h after CLP administration) were collected. 7.5 mL of full blood was transferred into collection systems containing ethylenediaminetetraacetic potassium salt (EDTA-K) (Sarstedt AG & Co., Germany) and immediately spiked with 37.5 μL of a 500 mM acetonitrile solution of 2-bromo-3-methoxyacetophenone (MPB). The addition of MPB is required for stabilization of reactive thiol H3 and H4 metabolites, as described by Takahashi et al. [18]. Approximately 1 mL of full blood was transferred into a clean polypropylene tube and stored at −25 °C until DNA isolation. Remaining aliquots of full blood were centrifuged for 10 min at 1620×g and separated plasma was stored at −25 °C until further analysis.
2.3 Genotyping
Genomic DNA was isolated from 100 μL of blood using commercially available kit according to manufacturer’s instruction (Blood Mini, A&A Biotechnology, Poland). CYP3A4*1G was identified by a PCR-restriction fragment length polymorphism method (RFLP). Reference sequence was obtained from Gene Bank (http://www.ncbi.nih.gov). The amplification cycles were performed in a T100 thermocycler (BIORAD, Singapore). Following settings were used: initial denaturation at 94 °C for 5 min, 37 cycles with denaturation at 94 °C for 30 s, annealing at 55 °C for 45 s and elongation at 72 °C at 30 s with subsequent final elongation at 72 °C for 7 min. Primers were designed in the Primer3Plus software [19] and sequences were as follows: F: 5′-CGTGGCCCAATCAATTATCT-3′, R: 5′-TTCTCCTGGGAAGTGGTGAG-3′. The reaction was performed in a volume of 25 μL with 1 μL of isolated DNA, 0.5 μM of each primer, 0.2 mM of each dNTP and 1 unit of DreamTaq polymerase in 1× PCR buffer (Thermo Scientific, Great Britain). MgCl2 was added to the reaction mix up to the final concentration of 2.5 mM. To detect the studied CYP3A4*1G polymorphism, the PCR product (220 bp) was digested with RsaI endonuclease (Roche Diagnostics, Germany). 15 μL of the product was incubated in 37 °C for 1 h in a mixture consisting of 1 U of the RsaI enzyme and 2.5 μL of 10× concentrated restriction buffer L, provided by the manufacturer. Water was added to make up a final volume of 25 μL. Digestion products were separated in a 3 % agarose gel. Two fragments (155 and 65 bp) were observed in wt/wt homozygotes, one fragment (220 bp) in *1G/*1G homozygotes and 3 fragments in heterozygotes. RFLP analysis results were confirmed by sequencing the PCR product from 10 randomly selected samples, on a 3130xl Genetic Analyzer (Applied Biosystems HITACHI, USA). In present study, CYP2C19 phenotype was also included as a potential covariate influencing CLP response in a multivariate analysis. A detailed study of the effect of CYP2C19 on CLP pharmacokinetics and methods for CYP2C19 phenotype evaluation were described previously by Karaźniewicz-Łada et al. [20]. Briefly, CYP2C19 phenotype was determined according to the presence of *1, *2 and *17 alleles in CYP219 gene. CYP2C19*2 was determined by a PCR–RFLP method, as described by Giusti et al. [15] while an allele-specific PCR was performed to analyze CYP2C19*17 as described by Sim et al. [21]. Patients with *1/*17 and *17/*17 diplotypes were classified as ultrarapid metabolizers (UM), *1/*1—extensive metabolizers (EM) and *1/*2 and *2/*17—intermediate metabolizers (IM).
2.4 Pharmacokinetic and Pharmacodynamic Analysis
Concentrations of CLP and its metabolites were determined by a validated HPLC–MS/MS method, described by Karaźniewicz-Łada et al. [22]. The analysis was performed on an Agilent 1200 chromatograph coupled with 6410B Triple Quad tandem mass spectrometer (Agilent Technologies, USA). CLP, H3 and H4 derivatives, CLPM and internal standard (piroxicam) were separated in a Zorbax Plus C18 column (Agilent Technologies, USA) at 40 °C. Mobile phase consisted of water and HPLC gradient-grade acetonitrile (Merck, Germany) acidified with 0.1 % of formic acid. Specific transitions for the analytes were monitored in a multiple reaction monitoring (MRM) mode. Sample preparation was as follows: 250 µL of plasma was spiked with 25 µL of an internal standard solution (1000 ng/mL piroxicam). Proteins were precipitated by adding 475 µL of acetonitrile. Subsequently, samples were centrifuged (22,570×g) and supernatant was filtrated through 0.45 µm Mini UniPrep filters with regenerated cellulose (Agilent Technologies, UK). The filtrate was evaporated in vacuum under 40 °C. Prior to injection the dry residue was reconstituted in 200 µL of mixture consisting of acetonitrile and water (50:50, v/v) with 0.1 % formic acid. The concentrations of analytes were calculated from calibration curves, which were analyzed prior to each analytical run according to European Medicines Agency guidelines for Bioanalytical Method Validation [23]. The quantification ranges were as follows: for CLP: 0.25–5 ng/mL; for bromomethoxy derivatives of H3 and H4: 0.25–50 ng/mL; for CLPM: 50–10,000 ng/mL. Pharmacokinetic parameters of the analytes were calculated by a non-compartmental method using WinNonlin 6.2 (Pharsight, USA). The area under the concentration–time curve (AUC0–t) was estimated by trapezoidal rule. Elimination half-life (t 1/2) was estimated from ln2/k, where k is the elimination rate constant calculated from terminal segment of log plasma concentration–time plot. Maximum concentration (C max) and time to reach C max (t max) were established directly from measured plasma concentrations.
Pharmacodynamic response was estimated by an impedance method (Multiplate analyzer; Roche Diagnostics). Samples for the assay were collected into hirudin-coated S-Monovette systems (Sarstedt AG&Co., Germany) 2–3 h after CLP administration. The ADP-induced platelet aggregation test was performed in accordance with manufacturer’s instruction. Platelet aggregation was quantified as the area under the curve (AUCaggr) of arbitrary units vs. time (AU·min).
2.5 Statistical Analysis
Shapiro–Wilk test was performed to establish normality of the data. The differences between normally distributed variables in two groups were determined with Student’s t test, otherwise Mann–Whitney U test was applied. Multivariate analysis was performed to estimate influence of CYP3A4*1G and other covariates on pharmacokinetic parameters of CLP and its metabolites and pharmacodynamic response. For calculating the Hardy–Weinberg equilibrium, an online calculation tool was used [24]. Otherwise, statistical calculations were performed in Statistica 10 software (StatSoft, USA). A p ≤ 0.05 was considered statistically significant.
3 Results
3.1 Study Group Characteristics
Data collected from all 82 patients were included in the study. Majority of patients were suffering from at least one co-morbidity, such as diabetes mellitus, hypertension or dyslipidemia. Beside CLP and aspirin most frequently coadministered drugs were statins, beta-adrenergic blocking agents (BB) and proton pump inhibitors (PPI). Detailed patients’ characteristics are described in Table 1.
3.2 Genotyping
Genotype distribution and allele frequencies of the CYP3A4*1G polymorphism are presented in Table 1. Studied SNP was in Hardy–Weinberg equilibrium. 84.1 % of patients were wt/wt homozygotes, while 15.9 % were heterozygotes. We did not observe any *1G/*1G homozygotes in studied population.
3.3 Pharmacokinetics and pharmacodynamics
Figure 1 illustrates mean pharmacokinetic profiles of CLP and each metabolite—CLPM, H3 and H4 in patients with wt/wt and wt/*1G genotypes. Concentrations of the main metabolite (CLPM) were quantifiable up to 24 h after CLP administration. However, the parent compound and thiol metabolites are rapidly eliminated and their concentrations were below level of quantitation in majority of samples collected 4 h after drug administration [25]. Pharmacokinetic profiles for studied analytes were similar between homozygotes and heterozygotes. There were no statistically significant differences in mean concentrations at measurement times for CLP and its metabolites (p values ranging from 0.293 to 0.922).
The comparison of calculated pharmacokinetic parameters of CLP and its metabolites is presented in Table 2. Due to very low concentrations of a parent drug and thiol metabolites, we were not able to detect the analytes in all collected samples. Therefore, patient numbers presented in Table 2 vary between groups. Performed statistical analysis showed, that the values of pharmacokinetic parameters were not significantly different.
In Fig. 2 is presented the pharmacodynamic effect of CLP depending on CYP3A4 genotype. Almost all studied patients were classified as responders to the CLP. Only one patient exhibited AUCagr above 486 AU*min which is considered as a threshold value for CLP resistance [26]. Median AUCagr for wt/wt genotype was lower than AUCagr for heterozygotes. However, the observed difference was not statistically significant (p = 0.484).
3.4 Multivariate Analysis
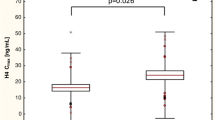
Multivariate analysis was performed to estimate, whether CYP3A4*1G allele might be an important covariate of CLP active metabolite pharmacokinetics and pharmacodynamics. We have investigated the influence of CYP3A4*1G on Cmax and AUC0–t of H3 and H4, and AUCagr. Following characteristics were examined as potential covariates: gender, age, BMI, diabetes mellitus, co-medications: statins, PPI, CCB, and CYP2C19 phenotype. Due to the fact that majority of patients were treated with statins, which vary widely in pharmacokinetic properties, we have focused on comparison of the type of statin (atorvastatin, simvastatin, rosuvastatin). To ensure normal distribution of the variables, values of pharmacokinetic parameters were logarithmically transformed. We have found that CYP3A4*1G remained a non-significant factor when it was included in a multivariate analysis with the covariates mentioned above. Noteworthy, neither patients’ characteristics nor coadministered drugs were significantly influencing pharmacokinetics of CLP and its metabolites. Only CYP2C19 phenotype had an impact on H4 C max and AUC0–t, although a model including both CYP3A4*1G and CYP2C19 phenotype was not statistically significant (Fig. 3). Mean C max and AUC0–t values of H4 active metabolite were similar in GG and GA carriers; however, there was a visible difference in the analyzed parameters between CYP2C19 phenotypes: UM, EM and IM. Detailed results of multivariate analysis are presented as supplementary material (Online Resource 1, 2 and 3).
Influence of CYP3A4*1G and CYP2C19 phenotype on C max (a) and AUC0–t (b) of H4 metabolite. Data are presented as means with 95 % confidence intervals as whiskers. Presented values of p refer to the model. UM CYP2C19 ultrarapid metabolizer, EM CYP2C19 extensive metabolizer, IM CYP2C19 intermediate metabolizer, AUC0–t area under the concentration time curve from time zero to time t, C max maximum plasma concentration, CYP cytochrome P450
4 Discussion
Effective antiplatelet treatment is particularly important in patients with a high risk of cardiovascular events. CLP is a widely used drug in prevention of thrombosis. However, occurrence of CLP resistance which has both non-genetic and genetic background diminishes effectiveness of the therapy. Polymorphisms of CYP2C19 are most widely studied among genetic factors. Presence of loss-of-function alleles, most notably *2 and *3, is associated with higher platelet reactivity and, therefore, higher risk of antiplatelet treatment failure [27, 28]. Influence of CYP3A4*1G on CLP pharmacokinetics and pharmacodynamics is unclear. Some authors notice, that the presence of CYP3A4*1G might be connected with lower activity of the enzyme [29, 30]. Yuan et al. [29] have recently found that CYP3A4*1G is associated with lower expression of CYP3A4 defined as mRNA concentration. The authors have also noticed a significant positive correlation between CYP3A4 mRNA and in vitro metabolic rate of fentanyl, which is predominantly metabolized by CYP3A4. Since CYP3A4 is one of the enzymes participating in bioactivation of CLP, lower concentrations of H3 and H4, as well as higher platelet reactivity might be observed in carriers of *1G allele. In our study, *1G allele frequency was similar in comparison to other studies that included European populations [14, 15]. We did not observe statistically significant difference in platelet reactivity. However, median AUCagr was higher in heterozygotes than in wild-type homozygotes. This finding is consistent with the results obtained by Fontana et al. [31], who did not find any association between CYP3A4*1G and platelet aggregation following administration of the same dose of CLP. However, it is contrasting with a study by Angiollilo et al. [14]. It showed that presence of polymorphic allele variant was associated with lower ADP-induced platelet aggregation. Above-mentioned studies focused only on the influence of CYP3A4 polymorphism on pharmacodynamic effect of the drug and did not include plasma concentrations of CLP and its metabolites. In the present study pharmacokinetic aspect of drug effectiveness was also considered. The results showed that in CYP3A4*1G carriers concentrations of CLP and its metabolites and values of calculated pharmacokinetic parameters were not significantly different as compared to wt/wt homozygotes (Fig. 1; Table 2).
CYP3A4 is responsible for transformation of almost 40 % of 2-oxo-CLP to the active thiol metabolite [6]. We investigated if *1G allele might influence CLP pharmacokinetics and pharmacodynamics in the presence of known CYP3A4 substrates (CCB, omeprazole and other PPI, atorvastatin and simvastatin). In an article by Park et al. [32], carriers of *1G allele were more vulnerable to occurrence of high-on-treatment platelet reactivity while being treated with CCB. In the present study, we did not observe a relationship between concomitant use of CCB, presence of *1G and antiplatelet effect of CLP and active metabolite pharmacokinetics. An impact of coadministration of PPI, most noteworthy omeprazole, on the efficacy of CLP therapy is most widely discussed. It was noticed that C max and AUC0–t of CLP active metabolite were lower and the platelet aggregation was higher while omeprazole or esomeprazole were administered [33, 34]. However, in our study after controlling for this covariate the difference in H3 and H4 pharmacokinetic parameters in *1G carriers remained not significantly different. Statins that are mostly metabolized by CYP3A4 might also influence lower rate of formation of H3 and H4. In present study, statistical analysis did not show a significant influence of statins and CYP3A4*1G on pharmacokinetics of CLP active metabolite in the study group.
The factors of largest influence on CLP pharmacokinetics and pharmacodynamics are CYP2C19 loss-of-function alleles. Park et al. [16] noticed that in CYP2C19 poor metabolizers CYP3A4*1G was related with lower value of P2Y12 reaction units. Authors suggest that *1G polymorphism might have an impact on CLP efficacy only in patients with severe CYP2C19 dysfunction. We have found that after including patients’ CYP2C19 phenotype into the model there was no statistically significant difference in AUCagr and pharmacokinetic parameters of H3 and H4. However, CYP2C19 phenotype remained significant predictor of H4 C max and AUC0–t. It is consistent with article presented by Giusti et al. [15]. The authors report no influence of CYP3A4*1G on ADP-induced aggregation, while CYP2C19*2, most common CYP2C19 loss-of-function allele, was associated with response variability. In conclusion, CYP3A4*1G may not be considered as an independent variable in CLP resistance. It has to be noted, however, that CYP3A4*1G is in the linkage disequilibrium with CYP3A4*1B which is linked with CYP3A5*1 (D′ = 0.757 between CYP3A4*1G and CYP3A4*1B, and D′ > 0.7 between CYP3A4*1B and CYP3A5*1) [14, 35]. Therefore, the presence of *1G might be only a marker of yet unstudied allele of greater influence of the variability on CLP therapy.
The study has several limitations. The study group is relatively small and ethnically homogenous. Moreover, observed concentrations of CLP and its thiol metabolites H3 and H4 were very low and in a number of samples were below quantitation limit of applied method. Therefore, full pharmacokinetic profiles could not be obtained for all patients. In addition, no *1G/*1G homozygotes were observed in the studied population and the results reflect only differences between wt/wt and wt/*1G genotypes. Presumably, a substantially larger sample size would be required to fully evaluate the gene effect.
5 Conclusions
The present study is a first to investigate the impact of CYP3A4*1G on clinical pharmacokinetics of CLP and its active metabolite. It was found that there is no difference in CLP pharmacokinetics as well as pharmacodynamics in carriers of *1G allele compared to wild-type homozygotes.
References
European Medicines Agency—Plavix—summary of product characteristics. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/000174/WC500042189.pdf. Accessed 14 Oct 2015.
Nguyen TA, Diodati JG, Pharand C. Resistance to clopidogrel: a review of the evidence. J Am Coll Cardiol. 2005;45:1157–64.
Mallouk N, Labruyère C, Reny J-L, Chapelle C, Piot M, Fontana P, et al. Prevalence of poor biological response to clopidogrel: a systematic review. Thromb Haemost. 2012;107:494–506.
Angiolillo DJ. Variability in responsiveness to oral antiplatelet therapy. Am J Cardiol. 2009;103:27A–34A.
Karaźniewicz-Łada M, Danielak D, Główka F. Genetic and non-genetic factors affecting the response to clopidogrel therapy. Expert Opin Pharmacother. 2012;13:663–83.
Kazui M, Nishiya Y, Ishizuka T, Hagihara K, Farid NA, Okazaki O, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010;38:92–9.
Tuffal G, Roy S, Lavisse M, Brasseur D, Schofield J, Delesque Touchard N, et al. An improved method for specific and quantitative determination of the clopidogrel active metabolite isomers in human plasma. Thromb Haemost. 2011;105:696–705.
Jang J-S, Cho K-I, Jin H-Y, Seo J-S, Yang T-H, Kim D-K, et al. Meta-analysis of cytochrome P450 2C19 polymorphism and risk of adverse clinical outcomes among coronary artery disease patients of different ethnic groups treated with clopidogrel. Am J Cardiol. 2012;110:502–8.
Mega JL, Simon T, Collet J-P, Anderson JL, Antman EM, Bliden K, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–30.
Yang W, Zhao D, Han S, Tian Z, Yan L, Zhao G, et al. CYP3A4*1G regulates CYP3A4 intron 10 enhancer and promoter activity in an allelic-dependent manner. Int J Clin Pharmacol Ther. 2015;53:647–57.
Yuan R, Zhang X, Deng Q, Wu Y, Xiang G. Impact of CYP3A4*1G polymorphism on metabolism of fentanyl in Chinese patients undergoing lower abdominal surgery. Clin Chim Acta. 2011;412:755–60.
He B-X, Shi L, Qiu J, Zeng X-H, Zhao S-J. The effect of CYP3A4*1G allele on the pharmacokinetics of atorvastatin in Chinese Han patients with coronary heart disease. J Clin Pharmacol. 2014;54:462–7.
Uesugi M, Hosokawa M, Shinke H, Hashimoto E, Takahashi T, Kawai T, et al. Influence of cytochrome P450 (CYP) 3A4*1G polymorphism on the pharmacokinetics of tacrolimus, probability of acute cellular rejection, and mRNA expression level of CYP3A5 rather than CYP3A4 in living-donor liver transplant patients. Biol Pharm Bull. 2013;36:1814–21.
Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Ramirez C, Cavallari U, Trabetti E, et al. Contribution of gene sequence variations of the hepatic cytochrome P450 3A4 enzyme to variability in individual responsiveness to clopidogrel. Arterioscler Thromb Vasc Biol. 2006;26:1895–900.
Giusti B, Gori AM, Marcucci R, Saracini C, Sestini I, Paniccia R, et al. Cytochrome P450 2C19 loss-of-function polymorphism, but not CYP3A4 IVS10+12G/A and P2Y12 T744C polymorphisms, is associated with response variability to dual antiplatelet treatment in high-risk vascular patients. Pharmacogenet Genomics. 2007;17:1057–64.
Park JJ, Park KW, Kang J, Jeon K-H, Kang S-H, Ahn HS, et al. Genetic determinants of clopidogrel responsiveness in Koreans treated with drug-eluting stents. Int J Cardiol. 2013;163:79–86.
Savcic M, Hauert J, Bachmann F, Wyld PJ, Geudelin B, Cariou R. Clopidogrel loading dose regimens: kinetic profile of pharmacodynamic response in healthy subjects. Semin Thromb Hemost. 1999;25(Suppl 2):15–9.
Takahashi M, Pang H, Kawabata K, Farid NA, Kurihara A. Quantitative determination of clopidogrel active metabolite in human plasma by LC–MS/MS. J Pharm Biomed Anal. 2008;48:1219–24.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115.
Karaźniewicz-Łada M, Danielak D, Rubiś B, Burchardt P, Oszkinis G, Główka F. The influence of genetic polymorphism of Cyp2c19 isoenzyme on the pharmacokinetics of clopidogrel and its metabolites in patients with cardiovascular diseases. J Clin Pharmacol. 2014;54:874–80.
Sim SC, Risinger C, Dahl M-L, Aklillu E, Christensen M, Bertilsson L, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–13.
Karaźniewicz-Łada M, Danielak D, Teżyk A, Zaba C, Tuffal G, Główka F. HPLC–MS/MS method for the simultaneous determination of clopidogrel, its carboxylic acid metabolite and derivatized isomers of thiol metabolite in clinical samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;911:105–12.
European Medicines Agency—Guideline on bioanalytical method validation Available from: http://www.ema.europa.eu/ema/index.jsp?curl=pages/includes/document/document_detail.jsp?webContentId=WC500109686&murl=menus/document_library/document_library.jsp&mid=WC0b01ac058009a3dc. Accessed 29 Jan 2016.
Rodriguez S, Gaunt TR, Day INM. Hardy–Weinberg equilibrium testing of biological ascertainment for mendelian randomization studies. Am J Epidemiol. 2008;169:505–14.
Karaźniewicz-Łada M, Danielak D, Burchardt P, Kruszyna L, Komosa A, Lesiak M, et al. Clinical pharmacokinetics of clopidogrel and its metabolites in patients with cardiovascular diseases. Clin Pharmacokinet. 2014;53:155–64.
Bonello L, Tantry US, Marcucci R, Blindt R, Angiolillo DJ, Becker R, et al. Consensus and future directions on the definition of high on-treatment platelet reactivity to adenosine diphosphate. J Am Coll Cardiol. 2010;56:919–33.
Hulot J-S, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–7.
Cresci S, Depta JP, Lenzini PA, Li AY, Lanfear DE, Province MA, et al. Cytochrome p450 gene variants, race, and mortality among clopidogrel-treated patients after acute myocardial infarction. Circ Cardiovasc Genet. 2014;7:277–86.
Yuan J-J, Hou J-K, Zhang W, Chang Y-Z, Li Z-S, Wang Z-Y, et al. CYP3A4*1G genetic polymorphism influences metabolism of fentanyl in human liver microsomes in Chinese patients. Pharmacology. 2015;96:55–60.
Zhang W, Chang Y-Z, Kan Q-C, Zhang L-R, Li Z-S, Lu H, et al. CYP3A4*1G genetic polymorphism influences CYP3A activity and response to fentanyl in Chinese gynecologic patients. Eur J Clin Pharmacol. 2010;66:61–6.
Fontana P, Hulot J. S, de Moerloose P, Gaussem P. Influence of CYP2C19 and CYP3A4 gene polymorphisms on clopidogrel responsiveness in healthy subjects. J Thromb Haemost. 2007;5:2153–5.
Park JJ, Park KW, Kang J, Jeon K-H, Kang S-H, Ahn HS, et al. CYP3A4 genetic status may be associated with increased vulnerability to the inhibitory effect of calcium-channel blockers on clopidogrel. Circ J. 2013;77:1289–96.
Angiolillo DJ, Gibson CM, Cheng S, Ollier C, Nicolas O, Bergougnan L, et al. Differential effects of omeprazole and pantoprazole on the pharmacodynamics and pharmacokinetics of clopidogrel in healthy subjects: randomized, placebo-controlled, crossover comparison studies. Clin Pharmacol Ther. 2011;89:65–74.
Frelinger AL 3rd, Lee RD, Mulford DJ, Wu J, Nudurupati S, Nigam A, et al. A randomized, 2-period, crossover design study to assess the effects of dexlansoprazole, lansoprazole, esomeprazole, and omeprazole on the steady-state pharmacokinetics and pharmacodynamics of clopidogrel in healthy volunteers. J Am Coll Cardiol. 2012;59:1304–11.
Plummer SJ, Conti DV, Paris PL, Curran AP, Casey G, Witte JS. CYP3A4 and CYP3A5 genotypes, haplotypes, and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:928–32.
Acknowledgments
Authors would like to thank Artur Teżyk from the Department of Forensic Medicine, Poznan University of Medical Sciences for assistance with HPLC–MS/MS analysis of plasma samples and Paweł Boruń from Institute of Human Genetics, Polish Academy of Sciences for assistance in primer designing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The study was supported by Polish Ministry of Science and Higher Education, Grant Number NN 405 419739 and Poznan University of Medical Sciences Grant Number 502-14-03306413-09628. The study protocol was approved by the local Ethical Committee at Poznan University of Medical Sciences. All performed procedures were in accordance with the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Conflict of interest
Authors declare no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Danielak, D., Karaźniewicz-Łada, M., Wiśniewska, K. et al. Impact of CYP3A4*1G Allele on Clinical Pharmacokinetics and Pharmacodynamics of Clopidogrel. Eur J Drug Metab Pharmacokinet 42, 99–107 (2017). https://doi.org/10.1007/s13318-016-0324-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13318-016-0324-7