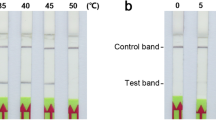
Abstract
Pantoea ananatis is a major pathogen that causes the new bacterial blight in rice, and its symptoms very similar to rice bacterial blight. Therefore, there is a dire need for an accurate and rapid method for detecting P. ananatis. In this study, an early and rapid visual detection method for P. ananatis was established. Using GyrB gene as the target sequence, an innovative recombinase-aided amplification detection system integrated with a lateral flow dipstick (RAA-LFD) was constructed. The optimized RAA-LFD detection method can be initiated at body temperature and does not rely on precise instruments. It does not require DNA extraction and can be used directly with plant tissue fluids. The results can be visualized after 10 minutes of amplification. The specificity and sensitivity tests showed that the RAA-LFD method could detect P. ananatis, whereas other common plant pathogens were not detected, and its detection sensitivity for P. ananatis DNA reached 100 copies/µL. The detection of diseased tissues indicated that this method could accurately detect P. ananatis in artificially inoculated rice tissues in the early stages of infection before symptoms. The RAA-LFD detection system established in this study is simple and fast, with visual results, excellent specificity, and high sensitivity. It is semi-quantitative and should be used for the early detection and rapid field diagnosis of new leaf blight, which provides technical support for the early warning and real-time detection of field samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Pantoea ananatis is a gram-negative bacterium which can be isolated from soil, water, plants, animals, human, etc. As a plant pathogenic bacteria, P.ananatis has many hosts, including pineapple, onion, eucalyptus, oyster mushroom, corn, wheat, rice, etc. The infections in different hosts perform various symptoms, such as rot of the stem, fruit and bulb, leaf spot disease and blight. It was reported that P.ananatis caused rice leaf blight in India, Benin, Togo and Russia. In 2020, P. ananatis was identified as the pathogen of a new rice bacterial blight. The blight has an incidence of 45%-60% in China. After the P.ananatis soaks the rice, water-soaked linear light brown lesions occur at the upper part of the leaves. Then the lesions spread down to the leaf margins, and the infected leaves turn light brown, eventually resulting in blade curling and wilting (Yu et al. 2022a, b).
The symptoms very similar to rice bacterial blight. From 2020 to 2021, this blight broke out in Zhejiang, Anhui, and other provinces in the middle and lower reaches of the Yangtze River in China, exhibiting an annually increasing trend. Yongyou, Zhongzheyou, Zhongzao, and other major varieties were all susceptible to this blight, whereas Thiamethoxam, Zinc-thiazole, Bismerthiazol, and other pesticides that are highly efficient for the preventive control of bacterial blight exerted insignificant effects on it (Yu et al. 2022a, b), which severely affected the safe production of rices and the sustainable development of this industry. Normally, early infected rices seedlings do not show symptoms; however, early diagnosis of blight is particularly important to avoid uncontrolled expansion. To conduct monitoring or preventive control, it is vital to rapidly and accurately detect low-level pathogens that are adequate for the early diagnosis of leaf blight. In practice, the timely detection of leaf blight at the early stages of infection will be conducive to making decisions to control this plant blight. Therefore, there is a need to develop early detection and preventive technologies for leaf blight.
The traditional technology of P. ananatis detection includes pathogen isolation and species identification based on morphological characteristics, which is difficult, time-consuming, and requires professional knowledge, which greatly increases the difficulty of detection (Teresa et al. 2006; Kini et al. 2019; Mamede et al. 2020; Mamede et al. 2018). Currently, the polymerase chain reaction (PCR) (Asselin et al. 2016; Shin et al. 2022; Figueiredo et al. 2012), multiplex PCR (Kini et al. 2021; Bangratz et al. 2020), qReal-Time PCR (qRT-PCR) (Shabnam et al. 2019), loop-mediated isothermal amplification (LAMP) (Kossi et al. 2021), and other molecular detection techniques have already been applied to the detection of pathogenic bacteria. PCR and qRT-PCR greatly improve the specificity and sensitivity of the detection, but their detection time is long, and their operations are complicated with demands for expensive materials and devices; hence, they are difficult to apply directly to the field (Huang and Yan 2020). LAMP detection exhibits strong specificity and simple operation; however, its primer design is complex and prone to generating false positives.
Recombinase-aided amplification (RAA) is an innovative technique for the rapid amplification of isothermal nucleic acids. It mainly consists of three core enzymes (recombinase, single-stranded DNA-binding protein, and DNA polymerase) undergoing enzymatic reactions at low temperatures to achieve rapid amplification of DNA or RNA, as a molecular biology detection technology (Lu et al. 2010). RAA shares similar principles as recombinase polymerase amplification (RPA). The difference is that the recombinases in the two reactions come from different origins. RPA recombinase originates from bacteriophage T4, and RAA recombinase is from bacteria or fungus. In recent years, the RAA technique has been widely used for the rapid detection of viruses, bacteria, mycoplasmas, parasites, and other species (Li et al. 2019; Xue et al. 2020a, b; Sun et al. 2020; Hu et al. 2019). The lateral flow dipstick (LFD) is an endpoint detection technology for visualizing the observation of amplification products. It is the product of biotin and carboxyl fluorescein (FAM) labels amplified by RAA, which applies antigen antibody binding to form "fluorescein antibody-double labeled nucleic acid amplification product-colloidal gold composite" at the detection line. The results can be directly observed with the naked eye. The RAA-LFD method integrating RAA and LFD detection technology presented strong specificity, high sensitivity, fast response, visualized results, and low standards for devices and operators and is applicable for quick checks in non-laboratory environments.
Currently, RAA technology, as a powerful monitoring tool, has been broadly applied in many fields, such as early monitoring and screening of pathogenic microorganisms, detection and identification of crop diseases and pests, rapid entry-exit quarantine, and food safety (Zhu et al. 2018; Ma et al. 2020; Fu et al. 2019; Li et al. 2019; Lai et al. 2022; Ju et al. 2020, 2022; Wang et al. 2021; Gao et al. 2022); however, there are no reports on the detection and application of P. ananatis in China or abroad. By forming the RAA-LFD detection system for P. ananatis, this study provides technical support not only for the rapid detection and identification of plant pathogens but also for further application in quick checks in basic-level quarantine departments and resource-limited laboratories.
Materials and methods
Strains for testing
Various plant pathogens including Pantoea ananatis, Xanthomonas oryzae pv. oryzae, Pantoea dispersa, Enterobacter cancerogenus, Xanthomonas oryzae pv. oryzicole, and Burkholderia glumae selected in this study are important pathogens of rice, including nine strains of P. ananatis and one strain of other types of pathogens. The strains of P. ananatis were provided by the Laboratory of Disease-Resistance and Breeding, whereas the other strains were provided by the Research Laboratory of Intelligent Forecasting Technology.
Sample preparation
DNA extraction with the kit: Lines were drawn on the Na Plate and cultured overnight at 37 °C, the ring scraper was inoculated to collect the bacterial solution, and the genomic DNA was extracted using a bacterial genomic DNA extraction kit following the manufacturer’s instructions.
Diseased leaves: Approximately 1 cm of diseased leaves were cut into pieces and placed in a 1.5 ml centrifuge tube. Sterile water (1 ml) was added, and the mixture was soaked for 10 min. The crude extract was used as a template. Bacterial solution: The bacterial solution was used directly as a template.
Instruments, devices, and reagents
Thermostatic metal bath, Test dipstick method by RAA amplification kit (RAA-nfo) (T00001, Jiangsu Qitian Gene Company), and LFD test dipstick (MGHD 1, Milenia Biotec, Germany).
Design of primers and probes
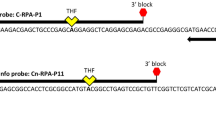
The GyrB gene encodes the DNA gyrase B subunit, which is a conserved housekeeping gene with a single copy. According to the gene sequence of P. ananatis GryB (GenBank No: MW981329.1) registered in the NCBI, DNAMAN software was used for sequence alignment, and Oligo software was applied to design primers and probes in the intraspecific conservative region. When integrated with the LFD, the 5' end of the downstream primer was labeled with biotin. The 5' end of the probe was labeled with the FAM fluorescent group, and the base 30 bp from the 5' end was replaced with Tetra-Hydro-Furan (THF). The detection primers and probes were synthesized by Shanghai Sangong Bioengineering Co. Ltd. (Shanghai, China).
Formation of RAA-LFD reaction system
The RAA reaction was performed using RAA-nfo kit following the manufacturer’s instructions. The total volume of the reaction system was 50 μL including 15.2 μL of double distilled water (ddH2O), 2.1 μL of each forward primer (10 μMol/L) and reverse primer (10 μMol/L), 0.6 μL of probe (10 μmol/L), 25.0 μL of basic buffer, 2.5 μL of DNA, all of which were added to the reaction tube after mixing to get dissolved fully, and finally, 2.5 μL of magnesium acetate was added to trigger the amplification reaction. The reaction conditions were maintained at constant temperature amplification of 39 °C for 20 min. The amplification product was diluted with buffer, and a 10 μL drop was placed on the sample pad. The LFD sample pad end was inserted vertically into a 100 μL dipstick expansion buffer (Milenia GenLine Dipstick Assay Buffer), and the reaction was continued at room temperature for 2 min. A positive LFD test dipstick shows both the quality control line and the detection line, indicating that the sample contains the nucleic acid fragment to be detected. A negative result indicates that the sample lacks the target nucleic acid fragment or has a quantity lower than the minimum detection quantity indicated by the test dipstick if only the quality control line appears.
Specificity test
Using P. ananatis as the test target, we selected the common rice pathogenic bacteria described in "Materials and methods" as the control group, the bacterial solution of the pathogenic bacteria as the template, the DNA of P. ananatis as the positive control, and sterilized ultra-pure water as the negative control, to test the specificity of the RAA-LFD reaction system.
Sensitivity test
Genomic DNA of P. ananatis was diluted by the 10-fold gradient dilution method to 10 ng·μL-1, 1 ng·μL-1, 100 pg·μL-1, 10 pg·μL-1, 1 pg·μL-1, and 100 fg·μL-1, with sterilized ultrapure water as the negative control, and the sensitivity of the reaction system to the target gene was tested.
The pure culture of P. ananatis was diluted at 105 CFU/mL in a 10-fold gradient, 1μL each was used as a template, and sterilized ultrapure water was used as a negative control to test the sensitivity of the formed reaction system to the pure culture of P. ananatis.
Usability test
To test the usability of the reaction system, diseased leaves from different regions of the Zhejiang Province were tested. A pure culture of P. ananatis was used as the positive control, and the sterilized ultrapure water was used as the negative control.
To test the effects of the reaction system on the detection of the diseased tissues, P. ananatis solution was prepared and inoculated on the rice leaves at the seedling stage. After activating the Pantoea ananatis strain, we cultured it overnight in PSA liquid medium with shaking. Then, we adjusted the inoculum concentration to an OD600 of 0.5 and inoculated rice leaves using the leaf-cutting method. Taking the tissues of the rice inoculation site, the template was prepared based on the method described in "Strains for testing", with a pure culture of P. ananatis as the positive control and sterilized ultrapure water as the negative control.
A bacterial suspension of 108 CFU/ml was inoculated to the rice using the leaf-cutting method, and water was used as the negative control. The samples were taken for examination 12h, 24h, 48h and 72h after the inoculation.
Analysis of applicable scope of detecting conditions
Amplification was performed at 33, 35, 37, 39, and 41 °C for 4, 8, 12, and 16 minutes, respectively. P. ananatis solution was used as the template, and sterilized water was used as the negative control. The developed RAA-LFD method was evaluated for its applicable temperature range and minimum amplification time, and the feasibility of temperature triggering amplification was also analyzed. we repeats three times for each RAA-LFD.
Results and analysis
Formation of RAA-LFD method for detecting Pantoea ananatis
Based on the principle of RAA primer design, four sets of primers (Table 1) were designed for the conserved region of the GryB gene of P. ananatis, which ranged from B1 to B5. The primers were screened using the RAA Basic Kit. The results showed that the five groups of primers could amplify a single specific band, but the blank control heterobands of P1, P2, P4, and P5 were bright (Fig. 1a). Therefore, the P3 primer pair was selected for validation. The probe for the paper dipstick method was designed manually and the P3 downstream primer with a biotin marker was synthesized again. The designed primer and probe combination were verified according to the RAA amplification system and procedures with P. ananatis solution as a template. The results showed that in the reaction system containing P. ananatis solution, there were two bands on the test dipstick: one in the quality control area and the other in the detection area. In the negative control, there was only one band in the quality control area, and there was no dipstick in the detection area (Fig. 1b). Furthermore, the amplified products were detected by agarose gel electrophoresis. The results indicated that the bands amplified by the designed primer pairs were single and clear, and the size of the target fragment was 165 bp (Fig. 1b). The results revealed that the RAA-LFD detection method for P. ananatis was preliminarily developed, and the combination of the designed primers and probes could be used for subsequent tests.
Evaluation of the specificity of RAA-LFD detection method for Pantoea ananatis
To verify the specificity of the combination of LFD-RPA primers and probes, RAA detection was performed on the tested P. ananatis strains and three common plant pathogens. After the reaction, the amplified product was detected using a flow cytometry dipstick. The results showed that the DNA and bacterial solution of P. ananatis pantomycetes were positive, with obvious detection lines in the detection area, whereas other pathogenic bacteria were negative, and no band was detected in the detection area (Fig. 2), indicating that the combination of RAA-LFD primers and probes designed in this study could specifically detect P. ananatis.
Sensitivity test of RAA-LFD method for detecting Pantoea ananatis
To validate the sensitivity of the RAA-LFD detection method, genomic DNA of P. ananatis diluted 10 times in the concentration gradient was used as the template for testing, and sterilized pure water was used as the negative control. Reference to the methods and standard curves of chen (2021). The results showed that all test dipsticks had visible control lines, and when the DNA concentration in the template was 107–102 copies/µL, the test dipstick presented a visible detection line (Fig. 3a), indicating that the minimum concentration of the RAA-LFD method for detecting genomic DNA of P. ananatis was 100 copies/µL.
The bacterial culture solution (105 CFU/mL) was diluted 10 times, and RAA-LFD detection was conducted on the diluted template. As shown in Fig. 3b, the band of the detection line tended to weaken with a decrease in the concentration of the bacterial solution. At a concentration of 102 CFU/mL, a visible detection line was still observed, indicating that the minimum concentration of the RAA-LFD method for detecting P. ananatis pure culture solution was 102 CFU/mL.
Usability test
The RAA-LFD primers designed in this study were used to amplify four diseased leaves from different regions of Zhejiang Jiangshu and Jiangxi provinces. The results showed that the LFD of the four diseased samples had quality control and detection lines (Fig. 4a), indicating that the RAA-LFD method has good universality. The RAA-LFD primers were used to amplify nine strains of P. ananatis, and the LFD of the nine strain samples had quality control and detection lines (Fig. 4b).
Utility test of the RAA-LFD method. a 1, diseased leaves from FuYang; 2, diseased leaves from Hangzhou; 3, diseased leaves from Zhenjiang; 4, diseased leaves from Jiangxi; 5, H2O. b, 9 strains of P. ananatis; c, the crude extract of rice tissue inoculated with P. ananatis for different symptoms; d, diseased leaves from different inoculation time
According to the method described in "Strains for testing", the crude extract of rice tissue inoculated with P. ananatis for different symptoms was used as a template for RAA amplification. LFD detection showed that the RAA-LFD method could accurately detect diseased tissues, indicating the incubation period of P. ananatis before symptoms appeared in the artificially inoculated rice tissues. These results were consistent with those of q-PCR (Fig. 4c).
According to the method in "Strains for testing", the rice tissues inoculated with P. ananatis at different times were cut into pieces, and the tissue solution was extracted as the template for RAA. The LFD detection exhibited that when the tissue solution template was the rice tissues inoculated for 12h-72h, the LFDs displayed quality control strips and detection strips. The color of the detection lines deepened with time, indicating that the amount of bacteria accumulated (Fig. 4d).
Determination of detecting condition range of RAA-LFD method
In this study, we developed a simple and rapid qualitative identification technology that can be used outside the laboratory. Therefore, the scope of application of the developed RAA-LFD method was evaluated, including the amplification temperature and amplification duration. The detection ability of the RAA-LFD method at different temperatures of 33, 35, 37, 39, and 41 °C, was individually determined using P. ananatis solution as a template. The temperature range screening results revealed that the primer pair had a good detection effect at 33–41 °C (Fig. 5a). To improve detection efficiency, the detection effect of the RAA-LFD method after amplification for 4, 8, 12, 16, and 20 minutes at 39 °C was determined. The amplification duration experiment showed a fuzzy detection line when the RAA-LFD reagent was amplified for 4 minutes and the detection line was visible when it was amplified for 8 minutes (Fig. 5b). Without any heating module, the staff could efficiently complete amplification and accurate detection by holding the PCR tube in the palm of their hand for 8 minutes (Fig. 5c).
Conclusions
The RAA-LFD detection system developed in this study was not only simple and fast, but also had visualized results, good specificity, and high sensitivity. It is applicable for early detection and rapid field diagnosis, providing technical support for the early warning of P. ananatis in rice and on-site detection of field samples.
Discussion
Novel rice leaf blight is a new type of bacterial disease that has emerged in Zhejiang, Anhui, and other provinces in the middle and lower reaches of the Yangtze River in recent years. This has caused huge losses in grain production, and the degree of its damage is increasing every year (Yu et al. 2022a, b; Teresa et al. 2006), which has severely affected the safe production of rice and sustainable development of this industry. Rapid and accurate detection of leaf blight pathogens in plants is key means to effectively preventing and controlling leaf blight. However, the existing pathogen identification methods can only be performed in the laboratory, and there is a delay between sample collection and result detection. We developed a visual detection method based on RAA and LFD detection. The crude extract of diseased tissue can be directly used as a template, which significantly reduces the template preparation time and improves detection efficiency. In contrast to the visualized loop-mediated-isothermal amplification (LAMP) detection system, which requires 60 minutes amplification at 65 °C, this method can complete the detection of samples in 10 minutes at 37 °C with visual results (Shabnam et al. 2019). This RAA-LFD method can be initiated even at human body temperatures. The entire detection process does not require any complex devices and is easy to operate, making its application beyond the laboratory environment to achieve the rapid detection of new rice leaf blight on site.
Usually, early infected rice plants do not exhibit any symptoms. Therefore, the early diagnosis of blight is particularly important to avoid uncontrolled blight expansion. The RAA-LFD method can accurately detect P. ananatis in artificially inoculated rice tissues at an early stage of infection before symptoms. Its detection result is consistent with that of fluorescent quantitative PCR; however, it does not require expensive devices or equipment and has a detection time of only 10 minutes. During detection using the RAA-LFD method, the DNA concentration of pathogenic bacteria is directly proportional to the color depth of the detection strip. The higher the DNA concentration, the deeper the color of the detection strips. Therefore, a colorimetric card displaying the color change of the detection strip can be prepared to establish the semi-quantitative rapid detection technology of RAA-LFD as a simple method for the real-time detection of pathogenic bacteria and rapid diagnosis of diseases.
Currently, the working principle of the LFD method for detecting RAA products is immuno-precipitation, and the primary cost involves the conjugated antibody on the dipstick. Reducing the quantity of combined monoclonal antibodies on the lateral flow dipstick could lower this cost. In addition, a microfluidic chip can be installed to replace the dipstick, which has already been successfully applied for rapid detection of SARS CoV-2 (Liu et al. 2021). At present, our team has been developing a filter paper dipstick method for extracting DNA and an isothermal amplification detection method based on a microfluidic paper-based chip with its supporting products and devices, which are expected to further reduce costs and improve efficiency.
Data Availability
All data included in this study are available upon request by contact with the corresponding author.
References
Asselin JE, Jean MB, Steven VB (2016) PCR Primers for Detection of Pantoea ananatis, Burkholderia spp., and Enterobacter sp. from Onion. Plant Dis 100(4):836–846
Bangratz M, Issa WK et al (2020) Design of a new multiplex PCR assay for rice pathogenic bacteria detection and its application to infer disease incidence and detect co-infection in rice fields in Burkina Faso. Plos One. https://doi.org/10.1371/journal.pone.0232115
Figueiredo JEF, Paccola-Meirelles LD (2012) Simple, Rapid and Accurate PCR-based detection of Pantoea ananatis in maize, sorghum and sdigitaria sp. J Plant Pathol 94(3):663–667
Fu MQ, Chen GF, Zhang CY et al (2019) Rapid and sensitive detection method for Karlodinium veneficum by recombinase polymerase amplification coupled with lateral flow dipstick. Harmful Algae 84:1–9
Gao B, Ma J, Li XH et al (2022) Development of RPA-LFD visualization assay for rapid detection of Ditylenchus destructor. Acta Phytopathol Sin. https://doi.org/10.13926/j.cnki.apps.000613
Hu JQ, Huang RN, Sun YT et al (2019) Sensitive and rapid visual detection of Salmonella typhimurium in milk based on recombinase polymerase amplification with lateral flow dipsticks. J Microbiol Methods 158:25–32
Huang WH, Yan MX (2020) Molecular detection of Phytophthora colocasiae of taro leaf blight based on PCR. J Landscape Res 12(1):33–35,38
Ju L, Jiang T, Aiying W et al (2022) A Rapid Detection Assay of Nilaparvata lugensBased on Recombinase Aided Amplification-lateral Flow Dipstick Technology. China J Rice Sci 36(1):96–104
Ju YL, Peng FS, Yan JF et al (2020) Development and application of Rc-RPA-LFD for the rapid detection of Rhizoctonia cerealis. Acta Phytopathol Sin 50(5):618–621
Kini K, Raoul A, Radiella D et al (2021) Genomics-Informed Multiplex PCR Scheme for Rapid Identification of Rice-Associated Bacteria of the Genus Pantoea. Plant Dis 105(9):2389–2394
Kini K, Dossa R, Dossou B et al (2019) A semi-selective medium to isolate and identify bacteria of the genus Pantoea. J General Plant Pathol 85(6):424–427
Kossi K, Issa W, Drissa S et al (2021) Development of two loop-mediated isothermal amplification (LAMP) genomics-informed diagnostic protocols for rapid detection of Pantoea species on rice. Methodsx 8:101216
Lai WH, Da RL, Bo D et al (2022) Development and Application of Recombinase-Aided Amplification Combined with Lateral Flow Dipstick Assay for Rapid Detection of Staphylococcus aureus. Food Sci 43(4):331–339
Li XN, Shen XX, Li MH et al (2019) Applicability of duplex real time and lateral flow strip reverse-transcription recombinase aided amplification assays for the detection of Enterovirus 71 and Coxsackievirus A16. Virol J 16(1):166
Li JL, Ma B, Fang JH et al (2019) Recombinase polymerase amplification (RPA) combined with lateral flow immunoassay for rapid detection of Salmonella in food. Foods 9(1):27
Liu D, Shen HC, Zhang YQ et al (2021) Amicrofluidic-integrated lateral flow recombinase polymerase amplification (MI-IF-RPA) assay for rapid COVID-19 detection. Lab on a Chip 10:2019–2026
Lu B, Cheng HR, Yan QF et al (2010) Recombinase-aid amplification: a novel technology of in vitro rapid nucleic acid amplification. Scientia Sinica Vitae 40:983–988
Ma B, Li JL, Chen K et al (2020) Multiplex recombinase polymerase amplification assay for the simultaneous detection of three foodborne pathogens in seafood. Foods 9(3):278
Mamede MC, Tebaldi ND (2020) Detection of Pantoea ananatis in corn seeds. Summa Phytopathologica 46(1):36–40
Mamede MC, Nilvanira DT, Lara CB et al (2018) Detection of Pantoea ananatis in corn seeds on semi-selective medium. Tropic Plant Pathol 43(3):254–256
Shabnam R, Sanni H, Renlin X et al (2019) A TaqMan real-time PCR for specific detection of Pantoea allii, a bacterial pathogen of onion. Plant Dis 103:3031–3040
Shin GY, Amy S, Teresa AC et al (2022) Validation of species-specific PCR assays for the detection of Pantoea ananatis, P. agglomerans, P. allii and P. stewartii. Plant Dis. https://doi.org/10.1094/PDIS-08-21-1810-SC
Sun XH, Hou Lai W, Li DR et al (2020) Research progress on the application of isothermal recombinase amplification in analytical detection. Food Ferment Industries 46(24):265–270
Teresa G, Stephanus N, Teresa A (2006) PA20, a semi-selective medium for isolation and enumeration of Pantoea ananatis J Microbiol Methods 64(2):225–231
Xue GH, Li SL, Zhang WW et al (2020) Reverse-transcription recombinase-aided amplification assay for rapid detection of the 2019 novel coronavirus (SARS-CoV-2). Anal Chem 92(14):9699–9705
Xue GH, Li SL, Zhao HQ et al (2020) Use of a rapid recombinase-aided amplification assay for Mycoplasma pneumoniae detection. BMC Infect Dis 20(1):79
Yu L, Yang C, Ji Y et al (2022) First Report of New Bacterial Leaf Blight of Rice Caused by Pantoea ananatis in Southeast China. Plant Dis 106(1):310
Yu L, Yang C, Ji Y et al (2022) Complete Genomic Data of Pantoea ananatis Strain TZ39 Associated with New Bacterial Blight of Rice in China. Plant Dis 106(2):751–753
Zhu P, Gao W, Huang H et al (2018) Rapid detection of Vibrio parahaemolyticus in shell fish by real-time recombinase polymerase amplification. Food Analytical Methods 11(8):2076–2084
Wang RB, Chen SZ, Zhao YM et al (2022) Development of a recombinase polymerase amplification-lateral flow dipstick (LFD-RPA) assay for rapid detection of Phytophthora colocasiae. J Plant Protect. https://doi.org/10.13802/j.cnki.zwbhxb.2021.2021178
Acknowledgments
The authors thank the disease-resistance rice breeding group (China National Rice Research Institute, China) for providing the strains.
Funding
“San Nong Jiu Fang”Sciences and Technologies Cooperation Project of Zhejiang Province (2022SNJF009); The Public Subject of State Key Laboratory of Rice Biology (NO. 20210303); Central Public-intererst Scientific Institution Basal Research Fund (NO. CPSIBRF-CNRRI-202123).
Author information
Authors and Affiliations
Contributions
Wang Aiying and Liu Shuhua conceived and designed the project. Wang Aiying conducted most of the experiments and wrote the manuscript. Luo Ju, Yang Baojun and, Tang Jian contributed part of the experiments and analyzed data. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interests
The authors declare that they have no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aiying, W., Ju, L., Cilin, W. et al. Establishment and application of the Recombinase-Aided Amplification-Lateral Flow Dipstick detection method for Pantoea ananatis on rice. Australasian Plant Pathol. 52, 283–291 (2023). https://doi.org/10.1007/s13313-023-00918-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-023-00918-8