Abstract
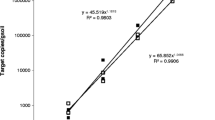
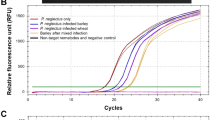
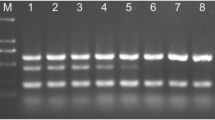
The soil-borne ascomycete Gaeumannomyces graminis var. tritici causes take-all of wheat (Triticum aestivum). Between host crops, G. graminis var. tritici survives saprophytically on crop debris and by infecting susceptible grass weeds or cereal volunteers. Invasion of roots in the following wheat crop results in reduced grain yield and quality. Take-all is commonly assessed in the field by visual inspection. Molecular-based methods are also available to detect G. graminis var. tritici, including a quantitative PCR (qPCR) assay that indirectly measures the amount of pathogen DNA in environmental samples. The qPCR is used as part of a commercial tool (known as PreDicta B™) to predict the risks of take-all in wheat crops prior to planting, which are dependent on the amount of Gaeumannomyces inoculum in field soils. Unfortunately, the costs associated with the PreDicta B™ test can be prohibitive to its use. As a result, in this study, an alternative qPCR assay was developed to measure directly the DNA of G. graminis var. tritici. The assay was shown to detect DNA of G. graminis var. tritici in both symptomatic and non-symptomatic wheat roots, with an increase in the amount of DNA detected having a strong relationship with an increase in take-all symptoms.



Similar content being viewed by others
References
Bithell SL, Butler RC, Harrow S, McKay A, Cromey MG (2011) Susceptibility to take-all of cereal and grass species, and their effects on pathogen inoculum. Ann Appl Biol 159:252–266
Bithell SL, McKay A, Butler RC, Herdina, Ophel-Keller K, Hartley D, Cromey MG (2012a) Predicting take-all severity in second-year wheat using soil DNA concentrations of Gaeumannomyces graminis var. tritici determined with qPCR. Plant Dis 96:443–451
Bithell SL, McKay A, Cromey MG (2012b) Low frequency of Gaeumannomyces graminis var. avenae in New Zealand; implications for take-all management in wheat. Australas Plant Pathol 41:173–178
Carbone I, Anderson JB, Kohn LM (1999) Patterns of descent in clonal lineages and their multilocus fingerprints are resolved with combined gene genealogies. Evolution 53:11–21
Cook RJ (2003) Take-all of wheat. Physiol Mol Plant Pathol 62:73–86
Herdina, Roget DK (2000) Prediction of take-all disease risk in field soils using a rapid and quantitative DNA soil assay. Plant Soil 227:87–98
Herdina, Harvey HP, Ophel-Keller K (1996) Quantification of Gaeumannomyces graminis var. tritici in infected roots and soil using slot-blot hybridization. Mycol Res 100:962–970
McMillan VE, Hammond-Kosack KE, Gutteridge RJ (2011) Evidence that wheat cultivars differ in their ability to build up inoculum of the take-all fungus, Gaeumannomyces graminis var. tritici, under a first wheat crop. Plant Pathol 60:200–206
Okubara PA, Schroeder KL, Paulitz TC (2005) Real-time polymerase chain reaction: applications to studies on soilborne pathogens. Can J Plant Pathol 27:300–313
Ophel-Keller K, McKay A, Hartley D, Herdina, Curran J (2008) Development of a routine DNA-based testing service for soilborne diseases in Australia. Australas Plant Pathol 37:243–253
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
White T, Bruns T, Lee S, Taylor J, Innis M, Gelfand D, Sninsky J (1990) PCR protocols: a guide to methods and applications. Academic, New York
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Acknowledgments
The authors thank Richard Falloon and Simon Bulman for their reviews of the manuscript. This work was funded by New Zealand’s Ministry for Business, Innovation and Employment through programme LINX0804.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Keenan, S., Cromey, M.G., Harrow, S.A. et al. Quantitative PCR to detect Gaeumannomyces graminis var. tritici in symptomatic and non-symptomatic wheat roots. Australasian Plant Pathol. 44, 591–597 (2015). https://doi.org/10.1007/s13313-015-0379-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-015-0379-y