Abstract
Introduction
Hypoglycemia remains a global burden and a limiting factor in the glycemic management of people with diabetes using basal insulins or oral antihyperglycemic drugs. Hypoglycemia data gleaned from randomized controlled trials (RCTs) have limited generalizability, as the strict RCT methodology and inclusion criteria do not fully reflect the real-world clinical picture. Therefore, real-world evidence, gathered from sources including electronic health records (EHR), is increasingly recognized as an important adjunct to RCTs.
Aims and methods
The LIGHTNING study applied advanced analytical methods, including machine learning (ML), to EHR data. The study aimed to predict hypoglycemic event rates in patients with type 2 diabetes (T2DM) receiving different basal insulin treatments to identify potential subgroups of patients who are at lower risk of hypoglycemia when treated with one basal insulin compared with another and to predict hypoglycemia-related cost savings in these subgroups. Here we provide an overview of the objectives, study design and methods, and validation approaches used in the LIGHTNING study.
Conclusion
It is hoped that results of the LIGHTNING study will help facilitate real-world clinical decision-making in addition to providing a clinically relevant predictive model of hypoglycemia risk.
Funding
Sanofi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hypoglycemia is a global burden and remains a limiting factor in achieving good glycemic control in patients with type 1 (T1DM) and type 2 diabetes mellitus (T2DM) [1]. Knowledge of hypoglycemia incidence with basal insulin (BI) use in general clinical practice is limited as the majority of data regarding hypoglycemia rates with BIs are derived from randomized controlled trials (RCTs). These RCT-derived hypoglycemia rates must be interpreted with caution; RCTs generally have strict inclusion and exclusion criteria, leading to exclusion of patients with complicating factors, e.g., those of older age with significant comorbidities, very poor glycemic control, and recurrent hypoglycemia, and therefore are not reflective of the true clinical picture.
While RCTs are considered the gold standard for evaluating the effects of drugs in specific disease and patient settings, it can be difficult to extrapolate results to more heterogenous populations in real-life clinical circumstances. Hypoglycemic event rates reported in RCTs are likely lower than in the real world [2], particularly for severe events [3]. In addition, hypoglycemic event rates are generally higher in observational studies versus RCTs [4], probably owing to the absence of patient selection, strict monitoring, education, and clinical oversight present in RCTs, and have been found to be influenced by different variables (e.g., geographic locations) [4].
Being able to identify people with different hypoglycemia risk profiles would enable optimal treatment management to be tailored toward these patient types. Real-world evidence (RWE) gathered from sources including electronic health records (EHRs), claims data, disease registries, and data from personal devices/software applications has the potential to be analyzed to establish these risk groups and factors associated with a greater number of hypoglycemic events. Indeed, information from RWE analyses is an important complementary component to clinical trial data as it provides a broader and unique insight into patient information, which could improve clinical decision-making. Thus, RWE derived from EHRs allows the generation of outcomes-based evidence from an unrestricted general diabetes population with a wide range of clinical phenotypes and comorbidities in routine clinical practice. However, it is necessary to overcome some of the limitations inherent in RWE collection and use. For example, data sources such as EHRs and claims data are not primarily generated or collected for research purposes (but rather to collect patient-centered information including clinical reports, health insurance, and reimbursement), and the lack of randomization and potential unobserved confounders could generate biased research outcomes [5].
Using advanced analytic approaches, such as machine learning (ML) [6], to analyze real-world data allows for hypothesis generation and modeling complex relationships/interactions. ML describes the programming of computers to "learn" complex data relationships, using training data or past experience and statistical theory [6], and capture them in a multivariate model. ML approaches can deal with large sets of variables, less-restrictive assumptions, inter-correlations among predicting factors, and automatic hypothesis generation. Leveraging rich structured (database) and unstructured (clinical notes) [7] data from EHRs, ML may be able to identify and model the hypoglycemia risk associated with BI use in patients with T2DM.
The LIGHTNING study involves the use of advanced analytical methods, including ML, to predict hypoglycemia rates in individuals with T2DM receiving different BI treatments. The study aims to identify general predictors of hypoglycemia in these patients, as well as potential subgroups of patients who are at lower risk of hypoglycemia with one type of BI formulation versus another, and estimate associated costs savings between comparison groups.
The approach to the LIGHTNING study is novel in terms of combining the latest ML methodology [utilizing natural language processing (NLP) to capture as many hypoglycemic events as possible] and rich real-world data sources to model hypoglycemia risk related to product use. Here, we provide an overview of the objectives, study design and methods, and validation approaches used in the LIGHTNING study.
The main objectives of the LIGHTNING study are to:
-
• Leverage ML-derived treatment-specific modeling to predict hypoglycemia rates in people with T2DM prescribed first- and second-generation BIs [first generation: insulin glargine 100 U/ml (Gla-100) and insulin detemir (IDet); second generation: insulin glargine 300 U/ml (Gla-300) and insulin degludec (IDeg)].
-
• Identify patient subgroups where the predicted hypoglycemic event rates differ between these BIs.
-
• Use an integrated claims–EHR data set to estimate incurred medical costs resulting from hypoglycemic events for these particular subgroups.
Methodology of the Lightning Study
Data Source
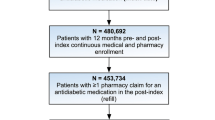
The Optum® longitudinal clinical repository (Humedica EHR) combines data from more than 50 US healthcare providers, covering more than 700 hospitals and 7000 clinics, in a database comprising more than 80 million patients. The "Integrated" data set links both claims and clinical data for approximately 10 million matched individuals. The LIGHTNING study will use all data collected from 1 January 2007 to 31 March 2017 from 831,456 people with T2DM receiving BI treatment (Fig. 1). All patient data from the Optum Humedica EHR and Integrated databases used for the LIGHTNING study were anonymized, and therefore informed consent from patients was not applicable.
LIGHTNING study population—patient and patient-treatment selection. a Multiple BI defined as patient-treatments that have another BI start within 1 week (before or after) of the specified BI start. b Inactivity defined as the lack of any time-stamped data. BI basal insulin, PSM propensity score matching
Optum’s Humedica EHR data sets were selected as the data source for the LIGHTNING study, owing to attributes including sample size, US geographic scope, richness of clinical data (especially clinical notes via NLP), and data quality. Optum had utilized NLP to identify and automatically read clinical documentation, translating it into structured data sets, which enabled the capture of key information from the notes, such as hypoglycemia occurrence [7].
Study Population
Inclusion Criteria
Patients with a confirmed diagnosis of T2DM [presence of 1 or more ICD-9 or 10 diagnosis codes (ICD-9: 250.x0; 250.x2; ICD-10: E11)] or one or more prescriptions for an antidiabetic treatment at any time during the study window were included. In addition, the cost estimation analysis required that patients had linked EHR and claims data available.
Exclusion Criteria
Patients who were likely to have a predominant diagnosis of T1DM at any time during the study window were excluded. Exclusion was based on the algorithm by Klompas et al. [8], which defined likely T1DM as a ratio of T1DM to T2DM diagnosis ICD codes > 0.5 and either a prescription for intramuscular glucagon or no record of an oral antidiabetic treatment other than metformin. Individuals who underwent more than ten BI treatments (i.e., frequent switching between BIs) within the study window were excluded, as they likely represent unusual clinical behavior.
"Patient-Treatment" and Cohort Creation
To maximize use of the available data, the LIGHTNING study aimed to evaluate data during time periods when people were using BI rather than simply evaluating each individual. Therefore, the unit of analysis used was the period in which an individual was observed to be receiving a BI treatment in the data set (termed a "patient-treatment"; Fig. 2); there could be multiple patient-treatments per individual.
The start of the patient-treatment (index date) was defined as the date of either the start of prescription of any BI (first generation: Gla-100 and IDet; second generation: Gla-300 and IDeg) or the change of prescription from one BI to another. The end of the patient-treatment was defined as the earliest of either the end of study period (31 March 2017), the change of prescription from the index BI to another, or 1 year after the treatment index date. For hypoglycemic event rate calculation, the duration was defined as the duration of the patient-treatment period minus the duration of all inpatient stays during this period (as inpatients may have been treated with BIs other than the index basal insulins, which may have contributed to the hypoglycemic events experienced).
Patients aged ≥ 18 years at the time of their first known prescription of a BI in the EHR database were analyzed. Patient-treatments were excluded from the analyses if more than one type of BI treatment was started within 1 week of the index date, if treatment started prior to 1 April 2015 (prior to Gla-300 becoming available in the USA), or if patients had any period of inactivity > 270 days during 1 year prior to index date.
The period prior to the patient-treatment (the "look-back" period) was used to collect data for covariate modeling. This period was 1 year by default, although covariates related to demographics and comorbidity may trace back to the beginning of the study period.
After patient and patient-treatment level inclusion/exclusion criteria had been applied, four treatment-specific modeling cohorts were created. Each modeling cohort represented all eligible treatment periods of the specific insulin and was used to develop a treatment-specific hypoglycemic model (the model that would be used for prediction of hypoglycemic event rates attributable to that particular BI; see "Modeling approach" section). The aggregate of the four cohorts—"BI-treated population"—were used as the scoring data set for all four final models.
Target Outcomes
Two primary outcomes were defined and identified: hypoglycemic event rates and hypoglycemia costs. Hypoglycemic event rates comprised both severe and non-severe events. An event was defined as hypoglycemia if it met any of the following four criteria: ICD-9 and 10 hypoglycemia diagnosis code (based on the algorithm of Ginde et al. [9]), laboratory plasma glucose levels ≤ 70 mg/dl, administration of intramuscular glucagon, or identified via NLP tables from the Optum EHR (Fig. 3). NLP is the method used to help define patient signs, disease, and symptoms from clinical notes. The NLP approach to identifying hypoglycemic events was based on the methodology utilized by Nunes et al. [7]. The search term used to define hypoglycemic events in the NLP table was “.*hypoglyce.*” excluding the terms “hypoglycemic awareness,” “hypoglycemic unawareness,” or “neonatal hypoglycemia.” The search also avoided inclusion of hits associated with "negative sentiment" (e.g., "negative" deny, have not) and identified hypoglycemia with sentiments that can be categorized as severe (e.g., severe, seizure, coma) or with any indication of historic occurrence (e.g., from patient histories). The definition of "severe" hypoglycemia is provided in Fig. 3; any hypoglycemic events that were not severe were defined as "non-severe."
Comprehensive definitions of hypoglycemia and severe hypoglycemia. a Maximum of one hypoglycemic event in a calendar day. In case of same-day hypoglycemic events, the severe event will be counted; secondary inpatient hypoglycemic events are excluded. b Codes used to identify hypoglycemia: ICD-9: 249.30; 249.80; 250.30; 250.31; 250.80; 250.81; 251.0; 251.1; 251.2; 270.3 (inclusion of 249.80, 250.80, and 250.81 only in the absence of other contributing diagnoses (ICD-9, 259.8, 272.7, 681.xx, 682.xx, 686.9x, 707.1–707.9, 709.3, 730.0–730.2, or 731.8). ICD-10: E08.64; E08.641; E08.649; E09.64; E09.641; E09.649; E10.64; E10.641; E10.649; E11.64; E11.641; E11.649; E13.64; E13.641; E13.649; E15; E16.0; E16.1; E16.2. c Codes regarded as severe by default: ICD-9: 249.30; 250.30; 250.31; 251.0; ICD-10: E08.641; E09.641; E10.641; E11.641; E13.641; E15. d ADA, EASD Joint Statement on Hypoglycemia [23]. ADA American Diabetes Association, EASD European Association for the Study of Diabetes, ED emergency department, ICD international classification of disease, IM intramuscular
Hypoglycemia costs were modeled utilizing the Integrated data set, with hypoglycemic events defined in the EHRs of adult T2DM patients who had both EHR and claims data, with costs captured from the claims data. Costs associated with hypoglycemic events were all medical costs incurred during the acute period between the start and the end of an event. The end of an event was defined as either (1) the discharge date of hospitalization (or 5 calendar days following admission if the discharge date was not available for inpatient admissions) or (2) the end of the next calendar day or the next hypoglycemic event for ED/outpatient visits (whichever occurred soonest).
Covariates for Modeling
The LIGHTNING study used both manually created covariates and covariates automatically created from all available data. These covariates were used for both the hypoglycemic rate model and cost-estimation analysis, although some covariates were excluded from cost analysis as it was not feasible to apply them to event-level data. The manually created covariates were identified through literature review as important confounding factors related to hypoglycemia and cost in T2DM, including: demographics, socioeconomics, comorbidities, diabetic complications, diabetic disease status, and medication use. Prior hypoglycemic event cost, only available in the claims database, was added to the cost-estimation analysis.
The automatically generated covariates were created by an algorithm that recognized patterns by clustering and re-grouping, as previously described [10]. Of the 1500 clusters identified in LIGHTNING, the top 100 clusters with the largest average number of patient-treatments in each variable were selected. The derived clusters were then evaluated for clinical relevance, as hierarchical clustering should be considered an intermediate step that requires validation.
Modeling Approach
Descriptive Analysis
Study variables, including key covariates and outcome measures, were analyzed descriptively, comparing the modeling cohorts. Numbers and percentages were reported for categorical variables and means and standard deviations (SDs) for continuous variables.
Hypoglycemia Prediction by Basal Insulin Type
To compare the hypoglycemic event rate between each first- or second-generation BI, a separate predictive model was developed for each treatment-specific cohort from the same list of covariates (Fig. 4). Once validated, each treatment-specific model was then applied to the full "BI-treated population" (scoring data set) to obtain the treatment-specific hypoglycemic event rate estimate (a prediction of the hypoglycemia rate in the full population if all patients were using that particular BI).
A Poisson generalized linear model (GLM) [11] was used for modeling because it allows a discrete count of events to occur in each period but does not allow for "over-dispersion" (i.e., it constrains the variance to be equal to the mean). A zero-inflated negative binomial GLM was initially used, but was over-fitted to the data for the smaller segments and performance was thus degraded. The Poisson model is also considerably simpler and so easier to understand. The Poisson GLM used the number of hypoglycemic events as the target variable (outcome) and the length of observation as an offset variable. Least absolute shrinkage and selection operator (LASSO) regularization [12] was used to select variables.
The models were developed ("trained") on 80% of each treatment-specific cohort (termed the "training sets"). Ten-fold cross-validation was used to inform model selection and model parameter optimization. Models were then validated on the remaining 20% of each treatment-specific cohort (internal validation). Bootstrapping was used to evaluate the variability of model estimates (i.e., to generate confidence intervals) [13].
Patient Subgroup Identification
Subgroups of the population with T2DM were identified through data-driven partitioning using all variables that had been selected by the LASSO regularization step. Categorical variables were represented as multiple binary variables, one for each level of the category. Numerical variables modeled as a continuum were, for the purposes of subgroup creation, split into quartiles or, in some cases, manually split if natural thresholds existed.
Within every partition or subgroup, the difference in hypoglycemia rates between BIs was assessed. To do this, the predictive hypoglycemia models developed for both BIs being compared ("reference" and "comparator") were applied to each patient-treatment in the scoring data set. Each patient-treatment was then associated with an expected "delta," e.g., the change in hypoglycemia rate that can be expected by changing from the comparator to reference BI. The average of these deltas across all patient-treatments within the subgroup constituted the differential rate for the subgroup. Ranking the subgroups by delta, one would identify the most differentiating subgroups between the comparator and reference BI, being the top and bottom subgroups.
Estimating Hypoglycemia Treatment Cost
A cost model was built to predict the cost of a hypoglycemic event in the T2DM population using the Integrated data set. The data set used for the treatment cost modeling included all hypoglycemic events in EHRs for T2DM patients who were at least 18 years of age at the time of the event and who had linked claims data. Severe hypoglycemic events with a cost of $0 were excluded. Study period and covariates were the same as those described for the hypoglycemia prediction models except when covariates could not be created because of data limitation.
Gradient-boosted trees (using prediction errors from previous decision trees to improve the performance of subsequent trees) were utilized for cost estimation, which have been successful in cost prediction previously [14]; they allowed the capture of complex non-linear relationships underlying the hypoglycemia cost. The cost estimator was applied to the subgroups identified as drivers of the differential hypoglycemic rate to estimate the cost per hypoglycemic event for each subgroup. When subgroups had key defining variables missing because of data limitation, an overall model cost estimate for a hypoglycemic event was used for the subgroups. Cost saving at the subgroup level was calculated by applying the subgroup-specific cost estimate of a hypoglycemic event to the delta hypoglycemic event rate between the comparator and reference BI.
Statistical Validation
Internal Validity
As previously mentioned, LIGHTNING used 20% of patient-treatments as the validation set for each treatment-specific model. This set was used to evaluate the model performance to ensure unbiased generalization if the model was applied to new data.
Overall validation of model accuracy was performed and reported using the normalized GINI coefficient, which was converted into an AUC equivalent measure. As part of the validation process, the statistical findings from the study were manually assessed for clinical relevance.
Determining the Confidence Interval
Bootstrapping [13] was used to determine the variability of the predictive models, estimating confidence intervals by running each model > 1000 times (each on resampling of the data) to observe the variability of estimates. This approach is an application to GLM of the non-parametric bootstrapping method as previously described [15,16,17] and is applied in many areas, including machine learning [18, 19], to quantify variability in estimates from many types of mathematical models.
The entire modeling process in LIGHTNING was bootstrapped, from sample selection through model training, to regularization, to calculation of the differential hypoglycemia rate estimates per subgroup. The bootstrapping process tested model predictions multiple times on the validation set, while different model parameters were tested; this process improves generality and confidence interval estimation (i.e., how confidently the results can be extrapolated from the analysis into broader populations).
Compliance with Ethics Guidelines
The OPTUM databases used in the LIGHTNING study were compliant with the Health Insurance Portability and Accountability Act. Fully anonymized retrospective data were obtained from OPTUM via a license agreement, and the LIGHTNING study did not involve primary data collection by the authors. The LIGHTNING study was therefore deemed exempt from ethical approval.
Discussion
The global burden of hypoglycemia is substantial, and significant gains could be made by identifying ways to reduce this burden with the use of particular therapies in certain patient subgroups. The LIGHTNING study aims to use the US Optum Humedica EHR and Integrated data sets to predict hypoglycemic event rates associated with use of first- and second-generation BI to identify subgroups of patients who benefit from a lower rate of hypoglycemia with one BI versus another and to estimate the cost savings within these subgroups to determine economic relevance to payors and health systems. ML-assisted predictive modeling techniques can exploit the rich data source provided, and NLP leverages the maximum amount of hypoglycemia data.
RWE data sources may have inherent bias that could limit their value for drawing causal inferences between treatment and outcomes. LIGHTNING aimed to minimize or avoid the impact of confounding factors that may impact the generalization of the modeling results by adopting an iterative, holistic approach combining data science, clinical expertise, and data quality control, which was undertaken to address potential biases (see Table 1 for examples of some of the methods used to address bias in the LIGHTNING study). Further strengths of the LIGHTNING study include the lack of restrictive inclusion criteria, which ensures that patients who may be at high risk of hypoglycemia are not excluded (as is often observed in RCTs); the inclusion of PSM, enabling comparisons with the DELIVER studies [20, 21] that used a different EHR data set (Predictive Health Intelligence Environment database); the capacity for the predictive modeling (and ML) techniques to handle large amounts of data; the hypothesis-free nature of the automatically generated covariates, which ensures that covariate determination is not limited by current clinical knowledge. However, there are some limitations to our methodology, which is to be expected when using novel techniques: results observed will not necessarily represent a causal relationship, but only statistical association (for example, covariates in the model may actually be proxies for other covariates not present in the database); the limited eligibility criteria used in the study may result in incomplete capture of clinical information from patients; the nature of the EHR means that only BI prescriptions are captured, not actual use of the product (or the dose used); the use of NLP is relatively new and should be further validated; the model used may be limited in its ability to capture the effects of interactions between covariates; the use of ML itself may have potential negative consequences, such as de-skilling of physicians due to overreliance on computer systems for decision-making [22]. In addition, it is important to note that external validation of the analyses using independent data sources (e.g., another EHR source) has not been done. Additional analyses with external data sets could provide a better understanding of relationships between variables and predicted outcomes and further validate the results. It should be noted that all study designs have limitations, and the limitations of LIGHTNING do not necessarily exceed those seen with more traditional methodologies.
In summary, the LIGHTNING study aims to provide RWE regarding hypoglycemic risks with different BIs for treatment of T2DM, leveraging ML algorithms and technology applied to the real-world data to identify patient subgroups that may experience a hypoglycemia benefit with newer BI formulations and to estimate the cost of hypoglycemia unique to that subgroup. It is hoped that findings generated from the LIGHTNING study, with oversight from a team of clinical experts, will support the development of novel clinical hypotheses in patients with T2DM that will help facilitate real-world clinical decision-making in addition to providing a clinically relevant predictive model.
References
Cryer PE. Hypoglycemia: still the limiting factor in the glycemic management of diabetes. Endocr Pract. 2008;14(6):750–6.
Elliott L, Fidler C, Ditchfield A, Stissing T. Hypoglycemia event rates: a comparison between real-world data and randomized controlled trial populations in insulin-treated diabetes. Diabetes Ther. 2016;7(1):45–60.
Pathak RD, Schroeder EB, Seaquist ER, Zeng C, Lafata JE, Thomas A, et al. Severe hypoglycemia requiring medical intervention in a large cohort of adults with diabetes receiving care in US Integrated health care delivery systems: 2005–2011. Diabetes Care. 2016;39(3):363–70.
Khunti K, Alsifri S, Aronson R, Cigrovski Berković M, Enters-Weijnen C, Forsén T, et al. Rates and predictors of hypoglycaemia in 27 585 people from 24 countries with insulin-treated type 1 and type 2 diabetes: the global HAT study. Diabetes Obes Metab. 2016;18(9):907–15.
Sherman RE, Anderson SA, Dal Pan GJ, Gray GW, Gross T, Hunter NL, et al. Real-world evidence—what is it and what can it tell us? N Engl J Med. 2016;375(23):2293–7.
Larranaga P, Calvo B, Santana R, Bielza C, Galdiano J, Inza I, et al. Machine learning in bioinformatics. Brief Bioinform. 2006;7(1):86–112.
Nunes AP, Yang J, Radican L, Engel SS, Kurtyka K, Tunceli K, et al. Assessing occurrence of hypoglycemia and its severity from electronic health records of patients with type 2 diabetes mellitus. Diabetes Res Clin Pract. 2016;121:192–203.
Klompas M, Eggleston E, McVetta J, Lazarus R, Li L, Platt R. Automated detection and classification of type 1 versus type 2 diabetes using electronic health record data. Diabetes Care. 2013;36(4):914–21.
Ginde AA, Blanc PG, Lieberman RM, Camargo CA Jr. Validation of ICD-9-CM coding algorithm for improved identification of hypoglycemia visits. BMC Endocr Disord. 2008;8:4.
Embrechts MJ, Gatti CJ, Linton J, Roysam B. Hierarchical clustering for large data sets. In: Georgieva P, Mihaylova L, Jain LC, editors. Advances in Intelligent Signal Processing and Data Mining: Theory and Applications. Berlin, Heidelberg: Springer; 2013. p. 197–233.
Nelder JA, Wedderburn RWM. Generalized linear models. J R Stat Soc Ser A (General). 1972;135(3):370–84.
Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B (Methodol) Wiley. 1996;58(1):267–88.
Mooney CZ, Duval RD. Bootstrapping: a nonparametric approach to statistical inference. Thousand Oaks: Sage Publications, Inc; 1993.
Guelman L. Gradient boosting trees for auto insurance loss cost modeling and prediction. Expert Syst Appl. 2012;39(3):3659–67.
DiCiccio TJ, Efron B. Bootstrap confidence intervals. Stat Sci. 1996;11(3):189–228.
Efron B. Nonparametric estimates of standard error: the jackknife, the bootstrap and other methods. Biometrika. 1981;68(3):589–99.
Efron B. Estimation and accuracy after model selection. J Am Stat Assoc. 2014;109(507):991–1007.
Wager S, Athey S. Estimation and inference of heterogeneous treatment effects using random forests. J Am Stat Assoc. 2017. https://doi.org/10.1080/01621459.2017.1319839.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Sullivan SD, Bailey TS, Roussel R, Zhou FL, Bosnyak Z, Preblick R, et al. Clinical outcomes in real-world patients with type 2 diabetes switching from first- to second-generation basal insulin analogues: comparative effectiveness of insulin glargine 300 units/ml and insulin degludec in the DELIVER D + cohort study. Diabetes Obes Metab. 2018;20(9):2148–58.
Zhou FL, Ye F, Berhanu P, Gupta VE, Gupta RA, Sung J, et al. Real-world evidence concerning clinical and economic outcomes of switching to insulin glargine 300 units/ml vs other basal insulins in patients with type 2 diabetes using basal insulin. Diabetes Obes Metab. 2018;20(5):1293–7.
Cabitza F, Rasoini R, Gensini GF. Unintended consequences of machine learning in medicine. JAMA. 2017;318(6):517–8.
International Hypoglycaemia Study Group. Glucose concentrations of less than 3.0 mmol/L (54 mg/dL) should be reported in clinical trials: a joint position statement of the american diabetes association and the european association for the study of diabetes. Diabetes Care. 2017;40(1):155–7.
Acknowledgments
Funding
This study and the article processing charges were funded by Sanofi. All authors had full access to all of the data in this study and take complete responsibility for the integrity of the data and accuracy of the data analysis.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole, and have given final approval to the version to be published.
Editorial Assistance
Editorial support for development of this manuscript was provided by Simon Rees, PhD, of Fishawack Communications, Ltd., supported by Sanofi.
Disclosures
Zsolt Bosnyak is an employee of Sanofi. Fang Liz Zhou is an employee of Sanofi. Javier Jimenez is an employee of Sanofi. Rachele Berria is an employee of Sanofi.
Compliance with Ethics Guidelines
All patient data from the Optum Humedica EHR and Integrated databases used for the LIGHTNING study were anonymized, and therefore informed consent from patients was not applicable.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Author information
Authors and Affiliations
Corresponding author
Additional information
Enhanced Digital Features
To view enhanced digital features for this article go to https://doi.org/10.6084/m9.figshare.7577747.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Bosnyak, Z., Zhou, F.L., Jimenez, J. et al. Predictive Modeling of Hypoglycemia Risk with Basal Insulin Use in Type 2 Diabetes: Use of Machine Learning in the LIGHTNING Study. Diabetes Ther 10, 605–615 (2019). https://doi.org/10.1007/s13300-019-0567-9
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13300-019-0567-9