Abstract
The B-cell activator factor (BAFF)/BAFF receptor (BAFF-R) axis seems to play an important role in the development and progression of chronic lymphocytic leukemia (CLL). Here, we investigated the association of eight single nucleotide polymorphisms (SNPs) in the BAFF (TNFSF13B) and BAFF-R (TNFRSF13C) genes with risk of sporadic CLL in a group of 439 CLL patients and 477 controls. We also examined the correlation between selected SNPs and CLL clinical parameters as well as BAFF plasma levels and intracellular BAFF expression. Our results point to a possible association between the rs9514828 (CT vs. CC + TT; OR = 0.74; CI 95 % = 0.57; 0.97; p = 0.022) and rs1041569 (AT vs. AA + TT; OR = 0.72; CI 95 % = 0.54; 0.95; p = 0.021) of BAFF gene and rs61756766 (CC vs. CT; OR = 2.03; CI 95 % = 1.03; 3.99; p = 0.03) of BAFF-R gene and CLL risk. Additionally, we observed that homozygotes rs1041569 AA and TT had a slightly higher risk (HR = 1.12) for the need of treatment in comparison to AT heterozygotes. In conclusion, our results indicate that SNPs in BAFF and BAFF-R genes may be considered as potential CLL risk factors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic lymphocytic leukemia (CLL) is one of the most prevalent leukemias in Western countries [1] with an incidence of 4.1 per 100,000 persons per year [2]. The characteristic feature of this disease is the gradual accumulation of mature B cells presenting typical markers such as CD5, CD19, CD20, and CD23 in lymphoid tissues, bone marrow, and peripheral blood (PB) [1, 3]. The CLL cell accumulation is caused by the disruption of programmed cell death rather than acute proliferation [1, 3].
B-cell activator factor (BAFF) (other names: BLys, TNFSF13B) and some other members of the tumor necrosis factor (TNF) family proteins have been shown to be engaged in providing survival signals to B cells by affecting genes associated with apoptosis [3]. BAFF binds to the following receptors: B cell maturation antigen (BCMA; TNFRSF17), transmembrane activator and calcium-modulator and cyclophilin ligand interactor (TACI; TNFRSF13B), and BAFF receptor (BAFF-R; TNFRSF13C) [4] which is a specific receptor for BAFF [5] and plays a key role in its biology [6].
BAFF-R is mainly expressed on B cells [6], and its expression on various B cell lines strongly correlates with BAFF binding to these cell lines [5]. Moreover, BAFF/BAFF-R signaling plays a central role in the survival and growth of normal and neoplastic B lymphocytes [5]. The aberrant expression of BAFF and BAFF-R has been reported, among others, in non-Hodgkin lymphomas (NHLs) including CLL [3, 5]. The SNP array designed to screen, inter alia, genes from TNF and TNF receptor superfamilies, as well as NFκB and related transcription factors, delineated BAFF (TNFSF13B) and BAFF-R (TNFRSF13C) genes as associated with NHL risk. Moreover, analysis conducted by NHL subtype revealed BAFF (TNFSF13B) to be associated specifically with CLL/small lymphocytic lymphoma (SLL) [7].
On the basis of these data, the hypothesis that the BAFF/BAFF-R pathway may play a role in the development and pathogenesis of this disease has been put forward [5].
The following observations support this hypothesis. Aberrant serum soluble BAFF levels have been reported in NHL patients including CLL patients [5, 8–11]. Correlations between BAFF expression and some clinical prognosis markers have been reported [5], and, what is more, the use of BAFF serum levels together with CD38, ZAP70 expression, and immunoglobulin heavy chain variable (IGHV) mutational status has been presented as a promising marker in CLL [12].
Genetic evidence also supports the above-mentioned hypothesis. Polymorphism rs9514828 (−871 C>T) in the promoter region of the BAFF gene has been associated with high levels of serum BAFF and familial CLL [3] and NHL risk [13]. A new mutation in the BAFF-R gene rs61756766 (His159Tyr) was determined in tumor and germline tissues from a subset of NHL patients [5, 6]. Additionally, the results of our preliminary study pointed to a possible association between genetic variations of BAFF and BAFF-R genes and the risk of sporadic CLL [14].
Since the BAFF/BAFF-R axis seems to play an important role in the development and progression of CLL, we decided to extend our previous study to investigate the association between single nucleotide polymorphisms (SNPs) of the BAFF and BAFF-R genes and CLL risk. We also examined the correlation between these SNPs and CLL clinical parameters as well as BAFF plasma level and intracellular BAFF protein expression.
Materials and methods
Study population
Patient population (N = 439) was composed of two cohorts of CLL patients. The majority of the first group (193 CLL patients; 85 females and 108 males) had entered into our previous study [14]. These patients were diagnosed with CLL in the Department of Hematology, Neoplastic Disease, and Bone Marrow Transplantation of Medical University of Wroclaw. The second group consisted of 246 patients (107 females and 139 males) diagnosed in the Department of Hematooncology and Bone Marrow Transplantation of Medical University of Lublin. Table 1 contains characteristics of CLL patients. Diagnosis of CLL was based on criteria from the International Workshop on Chronic Lymphocytic Leukemia (IWCLL) [15]. The follow-up period of this group was from 2 to 171 months (means ± SD = 58.01 ± 33.99). During this period, 106 patient indications for cytostatic treatment according to the IWCLL [15] were established. The period from the enrolment into the study and treatment ranged from 0.5 to 123 months (mean ± SD = 14.14 ± 21.75).
The control population was composed of 477 (296 subjects from our previous study and 181 additional subjects) randomly selected blood donors of Polish Caucasian origin (243 females and 234 males).
This study was approved by the Ethics Committee of the Medical University of Wroclaw and the Ethics Committee of the Medical University of Lublin. Written informed consent was obtained from all participants.
Selection of single nucleotide polymorphisms
SNPs, as previously described [14], were selected based on available literature [6, 16] and in silico analysis [17, 18].
The following SNPs of the 5′-untranslated region (UTR) of BAFF (TNFSF13B; 13q33.3) were examined: rs9514827 T>C (−2841); rs3759467 T>C (−2704); rs1041569 A>T (−2701); and rs9514828 C>T (−871) [14, 16]. According to in silico analysis, all these SNPs are located in the potential transcription factors binding sites (TFBS) [14, 17, 18].
As previously [14], we investigated four SNPs of BAFF-R (TNFRSF13C; 22q13.2): rs5996088 G>A (5′-near gene; potential TFB site); rs61756766 C>T (His159Tyr) [6] (Online Resource—Supplementary Fig. 1); rs7290134 T>C (3′-UTR; the potential miRNA binding site), and rs6002551 C>T (described by Wang et al. as a polymorphic variant of BAFF-R associated with NHL (7).
DNA isolation and genotyping
Genomic DNA was isolated from whole blood using Invisorb Blood Midi Kit (Stratec Molecular GmbH, Berlin, Germany) according to the manufacturer’s protocol. The following SNPs were genotyped by restriction fragment length polymorphism (RFLP): BAFF (rs1041569; rs9514827) [14, 16] and BAFF-R (rs61756766; rs6002551) [6, 14]. Primer sequences, annealing temperatures, and restriction enzymes are listed in Online Resource (Supplementary Table 1). Polymerase chain reactions (PCRs) were run on T100™ Thermal Cycler (Bio-Rad, Hercules, CA, USA). Allelic discrimination method with application of TaqMan SNP Genotyping Assays (Thermo Fisher Scientific, Waltham, MA, USA) was used to determine the following SNPs: BAFF (rs3759467, assay ID C_27497010_10) and BAFF-R (rs7290134, assay ID C_2189968_1_; rs5996088, assay ID C_30413471_10) [14]. The reactions were run on Applied Biosystems 7300 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA (Online Resource—Supplementary Table 2 contains a detailed list of assays used in this study). The BAFF rs9514828 was examined by both RFLP [16] (AciI, catalog no. R0551, New England Biolabs® Inc., Ipswich, MA, USA) and TaqMan methods (assay ID C_29641742_10) or by double genotyping with a TaqMan probe. Accuracy of genotyping methods for all SNPs was verified by direct sequencing of a few samples representing homozygotes of two types and heterozygotes for each investigated SNP. These samples were used as reference samples in the following genotyping experiments.
Intracellular analysis of BAFF
Peripheral blood samples from CLL patients were stained for intracellular BAFF expression as previously described [8]. Briefly, analysis of intracellular BAFF expression by CD19+ cells was performed on fresh PB samples. Monoclonal antibodies (MoAbs) used for analyses included anti-CD19 PE (Clone HIB 19, IgG1, BD Pharmingen, San Diego, CA, USA) and fluorescein isothiocyanate (FITC)-conjugated anti-BAFF (Clone 137314, IgG1, R&D Systems, Minneapolis, MN USA). The samples were analyzed by flow cytometry directly after preparation. For data acquisition and analysis, a FACSCalibur instrument with CellQuest software (Becton Dickinson, Franklin Lakes, NJ, USA) was used. The percentage of positive cells was measured from a cutoff set using an isotype-matched nonspecific control antibody.
Analysis of ZAP-70 expression in CLL cells
CLL cells were stained for ZAP-70 protein expression as previously described [8]. Briefly, ZAP-70-positive cells were evaluated in the PB via analysis of the surface expression of CD19 and CD5 antigens, as well as intracellular expression of ZAP-70 by flow cytometry. The percentage of CD19+CD5+ZAP-70+ cells was determined. MoAbs used for analyses included anti-ZAP-70 PE (Clone 1E7.2, IgG1, BD Biosciences, Franklin Lakes, NJ, USA) and anti-CD19 FITC (Clone HIB19, IgG1) and anti-CD5 PE-Cy5 (Clone UCHT2, IgG1) (BD Pharmingen, San Diego, CA, USA). The cutoff point for ZAP-70 positivity in leukemic cells was ≥20 %.
Detection of CD38 expression
As previously described [8], flow cytometry analysis of CD38 was performed on fresh PB samples stained with anti-CD38 FITC (Clone HIT2, IgG1), anti-CD5 PE-Cy5, and anti-CD19 PE (BD Pharmingen, San Diego, CA, USA). A standard, whole-blood assay with erythrocyte cell lysis was used for preparing the PB specimens. The samples were analyzed by flow cytometry directly after preparation. CLL cells were considered CD38-positive when ≥20 % was expressed in the membrane antigen.
Plasma BAFF immunoassay
As previously described [8], a commercial enzyme-linked immunosorbent assay (ELISA) kit, Quantikine Human BAFF/BLyS Immunoassay (R&D Systems, Inc. Minneapolis, MN, USA), was used for quantitative determination of human BAFF in plasma samples. We followed the protocol recommended by the manufacturer. The ELISA Reader Elx800 (BioTek Instruments, Winooski, VT, USA) was used [8].
Statistical analysis
As in our preliminary study [14], chi-square test, χ 2 df, was used to test the null hypothesis that cases and controls have the same distribution of genotype counts. To control type I error in the case of many tests for differences between SNP genotypes of cases and controls, a global (omnibus) chi-square test was performed, first to test the hypothesis zero, H0, which states that there were no differences between cases and controls in any SNP, opposite to the alternative, H1, which states that genotype frequencies in cases and controls were different at least in one SNP. Due to correlation between SNP distributions (linkage disequilibrium present), the distribution of global chi-square statistic was estimated numerically. In the case of small numbers, test statistics distribution was estimated numerically. Odds ratio (OR) was computed as the measure of effect size. Median was used as the location parameter. In the case of the median, the S n statistic was computed as the measure of variability: S n = med{med|x i − x j |; j = 1 ... n} [19]. S n is the typical distance between two randomly selected individuals and is used as the measure of variability instead of standard deviations when median is used instead of arithmetic mean. Additionally, the 1st and 3rd quartiles and minimal and maximal observations were reported. A linear model was used to test relations between SNP haplotypes and time to treatment (TTT) and requirement for introducing treatment. The likelihood ratio statistic, LRS ∼ χ 2, was used to test regression coefficients. The difference between two medians of plasma BAFF level was tested based on the bootstrapped studentized T statistic. Deviation from the Hardy-Weinberg equilibrium (HWE) was estimated with the chi-square test and measured as \( f=\frac{p_{CC}-{p}_C^2}{p_C\left(1-{p}_C\right)} \), where p C and p CC are allele C and genotype CC frequencies while f < 0 and f > 0 correspond to deficiency and excess of homozygotes, respectively, and f = 0 in the case of HWE. Haplotype frequencies (HFs) among SNPs were estimated with the maximum likelihood function [20]. Differences in genotype distributions between two groups of cases and between combined group of cases and controls were tested with the χ2 statistic, and the distribution of χ2 was estimated numerically. Analysis was performed using GNU Octave software version 3.8.2.
Results
The case-control study was carried out on groups of patients and controls described in the “Study population” section. There was no evidence that two groups of patients were different in terms of genotype distribution for any of the examined SNPs (χ 2 df≈39 = 43.97, p = 0.2673), so it allowed us to combine patients groups (193 + 246) in order to increase the statistical power of the present case-control study. The global (omnibus) test for homogeneity (χ 2 = 55 on approximately 35 degrees of freedom, ratio χ 2/df = 1.57) showed that the CLL group differed from the healthy control (HC) group in at least one SNP (χ 2 df=35 = 55; p = 0.026).
Hardy-Weinberg equilibrium
Seven out of eight SNPs investigated in this study were in the Hardy-Weinberg equilibrium in HC (Table 2 and Online Resource—Supplementary Table 3). The genotype distribution for rs9514828 differed from that expected under HWE in HC (p = 0.015; f = −0.116; CI 95 % = −0.21, −0.03). We did not exclude this variant from the analysis for several reasons. This particular SNP is functional and was the only one out of 20 SNPs examined (this manuscript presents only results for BAFF and BAFF-R) in our HC group for which deviation from HWE (DHW) was observed. It is well established that the most common sources of deviation from HWE are genotyping errors or population stratification [21]. Genotyping errors were excluded by double genotyping and by the fact that the patient group was in HWE. It is also hardly likely that our study population was stratified resulting from the admixture of other ethnic groups since both of the investigated groups are of Polish origin and they did not differ in terms of allele frequency. The C and T alleles frequencies were as follows: HC patients, C—56.9 %, T—43.1 %; and CLL patients, C—56.8 %, T—43.2 %.
From the statistical point of view, it is not surprising that one out of 20 SNPs tested at α = 0.05 was not in HWE, and as a matter of fact it is expected, even if all 20 examined SNPs in the general population were in HWE. Let s be the number of examined SNPs which are not in HWE in the sample at α = 0.05 and let r be the number of examined SNPs which are in HWE in the general population. We then get P(s ≥ 1| r = 20, α = 0.05) = 0.6415.
BAFF and BAFF-R polymorphisms and risk of CLL
In our preliminary study, we observed a difference in genotype distribution between the CLL patients and the controls for the rs9514828 BAFF variant (χ 2 df=1 = 3.946; p = 0.047) [14]. Here, in larger groups, we confirmed this difference (χ 2 df=1 = 5.23; p = 0.022) (Table 2). The risk of CLL for rs9514828 CT heterozygotes was lower than that for homozygotes CC (OR = 0.77; CI 95 % = 0.57, 1.03), while for rs9514828 TT homozygotes this risk was almost the same as for rs9514828 CC homozygotes (OR = 1.11 CI 95 % = 0.75, 1.63). Therefore, according to the parsimony rule, we assumed the overdominant model. The conducted analysis confirmed our assumption. In this model, heterozygotes had a protective effect (CT vs. CC + TT; χ 2 df=1 = 5.229; OR = 0.74; CI 95 % = 0.57, 0.97; p = 0.022).
In addition, we observed a difference in genotype distribution between the CLL patients and the controls for rs1041569 (χ 2 df=1 = 5.29; p = 0.02). Similarly, the genotype rs1041569 AT was protective when compared to rs1041569 AA (OR = 0.74; CI 95 % = 0.56, 0.99) (Table 2). The overdominant model applied in this case showed protective effect of heterozygote AT (AT vs. AA + TT; χ 2 df=1 = 5.886; OR = 0.72; CI 95 % = 0.54, 0.95; p = 0.021).
In our preliminary study [14], we observed a higher risk of CLL for rs61756766 CT heterozygotes, a very rare variant of the BAFF-R gene (OR = 1.79; CI 95 % = 0.73, 4.39), but the power of that study was low. Here, on larger groups, we noted a significant difference in genotype distribution between patients and controls (χ 2 df=1 = 4.43; p = 0.03) (Table 2). Moreover, we found the association between the rs61756766 CT genotype and the risk of CLL (OR = 2.03; CI 95 % = 1.03, 3.99). As previously [14], we did not determine any TT homozygote in patients or in controls.
Supplementary Table 3 (Online Resource) shows the distribution of the BAFF rs9514827 and rs3759467 and BAFF-R rs6002551, rs5996088, and rs7290134 gene polymorphisms investigated in this study for which distribution of genotypes was similar in patients and controls.
Additionally, we compared the distribution of BAFF and BAFF-R haplotypes between patients and controls. We noted a significant difference in haplotype distribution between HC and CLL subjects for BAFF-R (χ 2 df=6 = 17.4; p = 0.008) (Online Resource; Supplementary Table 4). There was no gene × gene interaction between BAFF and BAFF-R associated with the risk of CLL.
Plasma BAFF, intracellular expression of BAFF, and polymorphisms of BAFF
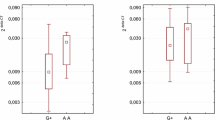
We investigated the possible association between all BAFF SNPs analyzed in this study as well as the (1) plasma BAFF level and the (2) intracellular expression of BAFF protein in PB CD19+ cells. We failed to find any of the BAFF SNPs examined in this study to be associated with plasma BAFF level (F df=8155 = 1.154, p = 0.331) (Online Resource; Supplementary Table 5) or to be associated with intracellular expression of BAFF in PB CD19+ cells (F df=8112 = 1.166, p = 0.326) (Online Resource; Supplementary Table 6).
BAFF and BAFF-R polymorphisms and clinical parameters
Since CD38 and ZAP70 are important prognostic markers of CLL, we divided patients into the following groups (using a 20 % cutoff value for CD38 and ZAP70): CD38+ and CD38−, and ZAP70+ and ZAP70−. Next, we compared the genotype distribution of all investigated SNPs of BAFF and BAFF-R between CD38+ and CD38− and ZAP70+ and ZAP70− CLL patients, but we did not find any significant differences. Also, none of the examined SNPs was associated with Rai stage.
Next, we analyzed correlations between the appearance of cytostatic treatment indications and time to treatment during the follow-up period of 2 to 171 months as well as the (1) haplotypes of BAFF and (2) rs1041569 and (3) rs9514828 of BAFF. There was no association between the requirement of the treatment and haplotypes (χ 2 df=8 = 4.38; p = 0.82) (Online Resource; Supplementary Table 7) as well as between treatment requirement and individual SNPs (rs1041569 χ 2 df=1 = 0.77; p = 0.68; rs9514828 χ 2 df=1 = 0.51; p = 0.48). Similarly, we did not observe a relationship between TTT and haplotypes of BAFF (Online Resource; Supplementary Table 8) (χ 2 df=6 = 5.97; p = 0.43).
Since we noticed that heterozygotes rs9514828 CT and rs1041569 AT had lower risk of CLL, in the next stage we analyzed TTT in relation to heterozygosity at rs1041569 and rs9514828 loci of the BAFF gene (Table 3). We noted that the average TTT for the rs1041569 AT heterozygotes was longer than in the group of AA and TT homozygotes. Homozygotes had 1.12 (HR = 1.12) times higher risk for the need of treatment (p = 0.316; CI 95 % = 0.7, 1.79). This result is encouraging because in practice it means that the TTT for patients carrying the AT genotype is more than two times longer than for patients carrying the AA or TT genotype (Fig. 1.) We also checked whether the heterozygotes at rs9514828 and rs1041569 would have longer overall survival (OS) in comparison to homozygotes, but we did not observe such a relation (p = 0.851 and p = 0.957, respectively).
Discussion
In our previous preliminary study, we investigated 20 SNPs of the BAFF/APRIL system and their association with CLL risk and some clinical parameters [14]. Here, we examined the association of BAFF and BAFF-R polymorphisms and CLL risk on a much larger group of patients (N = 439) and controls (N = 477) to confirm our previous findings [14].
The BAFF gene rs9514828 is located within the binding site of the myeloid zinc finger protein 1 (MZF1) transcription factor (TF) [9], and its functionality has been shown in luciferase assays, where the rs9514828 T allele was associated with higher luciferase activity [9, 22]. In contrast, however, Almeida and Petzl-Erler [23] who investigated BAFF expression by unstimulated cells (CD8+ T cells, monocytes, and NK cells) isolated from healthy individuals at protein and mRNA levels found that the rs9514828 CC genotype was associated with significantly higher BAFF expression.
The association of rs9514828 with different diseases such as primary Sjögren’s syndrome (pSS) [24], familial CLL [9], idiopathic thrombocytopenic purpura [25], hepatitis C virus-associated mixed cryoglobulinemia [26], pemphigus foliaceus [27], and T-cell lymphomas [22] has been reported. Of note, the protective or risk effect for C and T alleles has been shown for different diseases [9, 14, 22–25, 27].
In our previous study, we observed the difference in rs9514828 genotype distribution between CLL patients and HC (p = 0.047) [14].
Due to the above-mentioned contradictory data regarding the association of the rs9514828 variant with different diseases and the association of the T and C alleles with aberrant BAFF expression and our own data, we were prompted to verify these observations on larger groups of healthy and CLL subjects.
We confirmed our previous finding showing the difference in genotype distribution between CLL patients and controls for rs9514828. Based on obtained OR values, we assumed the overdominant model in which heterozygotes had protective effect.
As was mentioned earlier, rs9514828 is located within the MZF1-binding site [9]. We ran an in silico analysis using ConSite software [28] and found that the binding of MZF1 is predicted to be less strong for the C allele (score 8.510) than for the T allele (score 9.085). MZF1 has been shown to be involved in hematopoietic malignancies. MZF1 is a promoter/enhancer binding-type transcription factor and has been shown to act as a trans activator as well as a trans repressor, and it has been suggested that its oncogenic activity may be related to combined values of increased and decreased expression of target genes [29]. Moreover, ConSite software predicted the presence of a potential binding site for Sp1 transcription factor but only for the C allele (score 6.115). One of the mechanisms allowing neoplastic cells to survive and develop is the ability to overcome the intrinsic and extrinsic signals which in normal conditions lead to apoptosis. Sp1 transcription factor has been shown to be involved in regulation of numerous pro- and anti-apoptotic proteins such as BCL-2 or MCL1. It has also been reported that Sp1 regulation may act in two opposite directions. It may promote neoplastic cell resistance to apoptosis but may also promote these cells’ sensitivity to induction of apoptosis [30].
Taking into consideration the results of the in silico analysis, one may speculate that rs9514828 polymorphism may influence the regulation of the BAFF gene by affecting the binding sites of the above-mentioned transcription factors. The normal and neoplastic cells may differ in terms of expression level of these transcription factors which may influence the BAFF regulation.
Since both C and T alleles have been shown to have a different effect of BAFF expression in neoplastic cells, cell lines, and cells isolated from healthy donors and have been found to be predisposing or protecting in different diseases, it cannot be excluded that having both alleles may be advantageous by allowing cells to adjust to normal and/or abnormal conditions. Further research is required to test our hypothesis. First of all, it has to be investigated if the predicted in silico binding sites for transcription factors are functional. Subsequently, it has to be checked if and how these TFs regulate BAFF. Also, the expression level of MZF1 and Sp1 factors should be compared between CLL and normal cells.
According to our best knowledge, apart from our studies (present and preliminary [14]) in the literature there is only one other study investigating the association of BAFF gene polymorphism rs9514828 with sporadic CLL [10]. Novak et al. [10] did not find this variant to be associated with the risk of NHL or CLL. However, that study involved only 123 CLL/SLL cases [10]. Due to the discrepancy between this and our study, the association of rs9514828 with the risk of CLL remains to be examined in additional, independent studies. It needs to be clarified, whether this SNP is a true risk variant of CLL and if it is specific only for familial CLL [9] or both sporadic and familial CLL.
Apart from the association between rs9514828 and CLL, we also observed a significant difference in genotype distribution between CLL patients and controls for rs1041569. In the overdominant model, the AT heterozygotes showed a protective effect. Faustova et al. [31] reported a significant association of the rs1041569 T allele with myositis [31]. Nezos et al. [24] investigated rs1041569 polymorphism in primary Sjögren’s syndrome, but this group excluded this SNP from most of the conducted analysis since it was not in HWE in the healthy control group [24]. Recently, Lin et al. [32] have investigated rs1041569 in autoimmune thyroid diseases (Grave’s disease and Hashimoto’s thyroiditis). They did not find an association between this SNP and the examined diseases, but OR and CI 95 % reported by this group for rs1041569 AT heterozygotes (OR = 0.79; CI 95 % 0.57, 1.10) [32] were similar to the figure observed by us (OR = 0.74; CI 95 % = 0.57, 0.97). It is difficult to draw any definite conclusion regarding the ORs for TT homozygotes based on our and Lin et al.’s [32] studies due to the low frequency of this genotype observed in both populations as well as the CI 95 % values (our study, 2.3 %; CI 95 % = 0.96, 4.13; Lin et al.’s study [32], 1.1 %; CI 95 % = 0.14, 2.93).
The in silico analysis via application of ConSite [28] and SNPinfo [17] showed that this variant is located within the E47 (Thing1/E47) TFBS. The calculated matrix similarity for E47 (Thing1/E47) at position rs1041569 suggests better binding of this TF to the A allele (ConSite scores for A allele 9.982 and 8.897 for T allele; SNPinfo: A allele core similarity 1 and matrix similarity 0.928; T allele core similarity 1 and matrix similarity 0.911).
E12 and E47 are products of the alternative splicing of a single TCF3 (E2A) gene. They belong to the class I helix-loop helix (bHLH) proteins and are broadly expressed, multifunctional TFs playing a role in many developmental processes [33, 34]. These two proteins can act both as tumor suppressors and as tumor promoters [35]. E2A (E47) TF plays a key role in B-cell development, maturation, and function and regulates proliferation and survival of these cells [36].
It has been shown that E2A (E47) is overexpressed in CLL cells. Elimination of this TF caused increased apoptosis [36]. Additionally, ConSite prediction showed that the T allele caused the disappearance of the GATA-2 binding site (score 4.174 for A allele) and introduced the possible binding site for PU.1 (SPI-1 score 4.776). However, due to limited information available about the function of this SNP, it is difficult to propose the mechanism which will explain the observed protective effect for AT heterozygotes. As was mentioned above, the rs1041569 TT homozygotes are rare, and study on larger sample size of CLL patients and healthy controls is needed to verify the effect associated with this genotype and the risk of CLL. Moreover, our study is the first (according to our best knowledge) to report an association between rs1041569 and the risk of CLL, and as was mentioned above, further case-control studies on large groups are necessary.
The different softwares designed to predict potential TFBS provide results which have to be evaluated in functional studies. Therefore, to resolve all arisen discrepancies, further in silico, genetic and functional studies are needed.
As described above, we observed that heterozygosity at rs9514828 and rs1041569 loci of the BAFF gene may protect from CLL development. Therefore, we checked if rs9514828 CT and rs1041569 AT heterozygotes would have a better prognosis in terms of overall survival, time to treatment, or the requirement for introducing treatment. Neither rs9514828 CT nor rs1041569 AT heterozygotes had longer overall survival in comparison to both types of homozygotes. There was also no association between the requirement for treatment and heterozygosity at rs9514828 or at rs1041569. No association was found between time to treatment and heterozygosity at rs1041569. However, we observed that the average time to beginning of treatment was much longer for rs1041569 AT heterozygotes in relation to homozygotes AA and TT. This result is in agreement with the lower risk of CLL for rs1041569 AT heterozygotes, observed here. We are not able to provide an explanation of this observation at this moment, but the influence from interactions between TFs and administered drugs cannot be excluded.
It has to be mentioned that we possessed a limited number of samples with TTT data available. It will be interesting to test our hypothesis of a protective role of heterozygosity at rs9514828 and rs1041569 variants of the BAFF gene on a larger group of CLL patients and HC.
As all BAFF SNPs analyzed in this study are located in potential TFBS and the relationship between rs9514828 polymorphism and expression level of BAFF was shown in luciferase assays [9, 22] and with serum BAFF levels (sBAFF) [9], we also addressed these issues in our research. We did not find any of the SNPs of the BAFF promoter investigated in the present study to be associated with plasma BAFF levels or with intracellular expression of BAFF protein in PB CD19+ cells. Similarly, Ansell et al. [37] did not find any significant association between rs9514828 and sBAFF levels in follicular grade 1 non-Hodgkin lymphoma [37]. The same result was obtained for pSS [16]. We examined the same SNPs of the BAFF promoter region (rs9514827, rs3759467, rs1041569, rs9514828) as had been previously studied by Eilertsen [38] and Fabris [39] and colleagues, who had investigated these SNPs in the systemic lupus erythematosus and rheumatoid arthritis, respectively. These authors also failed to find a relationship between serum BAFF levels and investigated promoter variants of the BAFF gene [38, 39]. Previously, Novak et al. [9] found that sBAFF levels were higher in patients with familial CLL than with sporadic CLL and suggested the correlation between elevated levels of serum BAFF and the presence of a T at rs9514828 in the BAFF promoter [9]. Since our cohort probably did not contain familial cases, we did not observe such a correlation in our study.
In silico analysis as well as previously mentioned results showing the association of rs9514828 with aberrant expression of BAFF gene suggested that we and other authors should observe a correlation between at least this variant and the level of BAFF. Due to the data available, we investigated only the correlation between promoter variants and BAFF protein levels. Any additional studies have to be designed to more deeply examine the role of SNPs investigated here in the regulation of the BAFF gene.
BAFF-R is the main receptor for BAFF, and the BAFF/BAFF-R pathway is crucial for the survival and growth of CLL cells [5]. Hildebrand et al. [6] described an association between the BAFF-R rs61756766 and the risk of non-Hodgkin lymphoma such as the diffuse large B cell lymphoma, follicular lymphoma, lymphoplasmacytic lymphoma, and mucosa-associated lymphoid tissue, but not with CLL. In the same publication, they also reported a correlation between this variant of the BAFF-R gene and the increased recruitment of TRAF 2, 3, and 6 and showed that signaling through this variant of BAFF-R resulted in increased NF-κB1 and NF-κB2 activity [6]. In our study, we found that rs61756766 CT heterozygotes had two times higher risk of CLL than CC homozygotes. Of note, we genotyped altogether 916 subjects (patients and controls) and did not find any rs61756766 TT homozygote. The frequency of the T allele was 2.7 % in patients and 1.4 % in HC. The absence of TT homozygotes was expected, since the probability that there will be no TT homozygotes within 439 cases and 477 controls given the above-mentioned T allele frequencies is p = 0.6592. Due to such low frequency of patients carrying the CT genotype for whom complete clinical data were available, we were not able to investigate potential relations between the presence of this allele and clinical features. Apart from rs61756766, we genotyped three additional SNPs of the BAFF-R gene: rs6002551, rs5996088, and rs7290134 [14]. We did not find any of these SNPs to be associated with the risk of CLL, but we observed a significant difference in haplotype distribution (formed by four SNPs of BAFF-R) between CLL and HC patients, which confirms that genetic predisposition to CLL may be associated with the BAFF-R gene.
In conclusion, in this study we investigated genetic variations in BAFF and BAFF-R in CLL including the assessment of haplotypes, gene × gene interaction, and correlation with clinical features. Our case-control study indicates a possible association of the most widely studied rs9514828 SNP of the BAFF gene as well as, described here for the first time, the possible association of rs1041569 of the BAFF gene with the risk of CLL. Moreover, this is the first study which showed the association between the BAFF-R gene rs61756766 and CLL risk. Taking into consideration the fact that genetic predisposition to CLL is still not well established, our results may help to further investigate this issue and may ultimately help to establish which of the genetic variations reported in the literature are the true CLL risk factors.
References
Haiat S, Billard C, Quiney C, Ajchenbaum-Cymbalista F, Kolb JP. Role of BAFF and APRIL in human B-cell chronic lymphocytic leukaemia. Immunology. 2006;118(3):281–92.
Pflug N, Bahlo J, Shanafelt TD, Eichhorst BF, Bergmann MA, Elter T, et al. Development of a comprehensive prognostic index for patients with chronic lymphocytic leukemia. Blood. 2014;124(1):49–62.
Ferrer G, Hodgson K, Montserrat E, Moreno C. B cell activator factor and a proliferation-inducing ligand at the cross-road of chronic lymphocytic leukemia and autoimmunity. Leuk Lymphoma. 2009;50(7):1075–82.
Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9(7):491–502.
Yang S, Li JY, Xu W. Role of BAFF/BAFF-R axis in B-cell non-Hodgkin lymphoma. Crit Rev Oncol Hematol. 2014;91(2):113–22.
Hildebrand JM, Luo Z, Manske MK, Price-Troska T, Ziesmer SC, Lin W, et al. A BAFF-R mutation associated with non-Hodgkin lymphoma alters TRAF recruitment and reveals new insights into BAFF-R signaling. J Exp Med. 2010;207(12):2569–79.
Wang SS, Purdue MP, Cerhan JR, Zheng T, Menashe I, Armstrong BK, et al. Common gene variants in the tumor necrosis factor (TNF) and TNF receptor superfamilies and NF-kB transcription factors and non-Hodgkin lymphoma risk. PLoS One. 2009;4(4):e5360.
Bojarska-Junak A, Hus I, Chocholska S, Wasik-Szczepanek E, Sieklucka M, Dmoszynska A, et al. BAFF and APRIL expression in B-cell chronic lymphocytic leukemia: correlation with biological and clinical features. Leuk Res. 2009;33(10):1319–27.
Novak AJ, Grote DM, Ziesmer SC, Kline MP, Manske MK, Slager S, et al. Elevated serum B-lymphocyte stimulator levels in patients with familial lymphoproliferative disorders. J Clin Oncol Off J Am Soc Clin Oncol. 2006;24(6):983–7.
Novak AJ, Slager SL, Fredericksen ZS, Wang AH, Manske MM, Ziesmer S, et al. Genetic variation in B-cell-activating factor is associated with an increased risk of developing B-cell non-Hodgkin lymphoma. Cancer Res. 2009;69(10):4217–24.
Molica S, Digiesi G, Battaglia C, Cutrona G, Antenucci A, Molica M, et al. Baff serum level predicts time to first treatment in early chronic lymphocytic leukemia. Eur J Haematol. 2010;85(4):314–20.
Vincent FB, Saulep-Easton D, Figgett WA, Fairfax KA, Mackay F. The BAFF/APRIL system: emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev. 2013;24(3):203–15.
Lahiri A, Pochard P, Le Pottier L, Tobon GJ, Bendaoud B, Youinou P, et al. The complexity of the BAFF TNF-family members: implications for autoimmunity. J Autoimmun. 2012;39(3):189–98.
Jasek M, Wagner M, Sobczynski M, Wolowiec D, Kuliczkowski K, Woszczyk D, et al. Polymorphisms in genes of the BAFF/APRIL system may constitute risk factors of B-CLL—a preliminary study on a Polish population. Tissue Antigens. 2015;86(4):279–84.
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–56.
Nossent JC, Lester S, Zahra D, Mackay CR, Rischmueller M. Polymorphism in the 5’ regulatory region of the B-lymphocyte activating factor gene is associated with the Ro/La autoantibody response and serum BAFF levels in primary Sjogren’s syndrome. Rheumatology. 2008;47(9):1311–6.
Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37 :W600–5.Web Server issue
Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34 :W635–41.Web Server issue
Rousseeuw PJ, Croux C. Alternatives to the median absolute deviation. JASA. 1993;88:1273–83.
Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12(5):921–7.
Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet: EJHG. 2005;13(7):840–8.
Zhai K, Tian X, Wu C, Lu N, Chang J, Huang L, et al. Cytokine BAFF gene variation is associated with survival of patients with T-cell lymphomas. Clin Cancer Res: Off J Am Assoc Cancer Res. 2012;18(8):2250–6.
de Almeida ER, Petzl-Erler ML. Expression of genes involved in susceptibility to multifactorial autoimmune diseases: estimating genotype effects. Int J Immunogenet. 2013;40(3):178–85.
Nezos A, Papageorgiou A, Fragoulis G, Ioakeimidis D, Koutsilieris M, Tzioufas AG, et al. B-cell activating factor genetic variants in lymphomagenesis associated with primary Sjogren’s syndrome. J Autoimmun. 2014;51:89–98.
Emmerich F, Bal G, Barakat A, Milz J, Muhle C, Martinez-Gamboa L, et al. High-level serum B-cell activating factor and promoter polymorphisms in patients with idiopathic thrombocytopenic purpura. Br J Haematol. 2007;136(2):309–14.
Gragnani L, Piluso A, Giannini C, Caini P, Fognani E, Monti M, et al. Genetic determinants in hepatitis C virus-associated mixed cryoglobulinemia: role of polymorphic variants of BAFF promoter and Fcgamma receptors. Arthritis Rheum. 2011;63(5):1446–51.
Malheiros D, Petzl-Erler ML. Individual and epistatic effects of genetic polymorphisms of B-cell co-stimulatory molecules on susceptibility to pemphigus foliaceus. Genes Immun. 2009;10(6):547–58.
Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32 :W249–52.Web Server issue
Eguchi T, Prince T, Wegiel B, Calderwood SK. Role and regulation of myeloid zinc finger protein 1 in cancer. J Cell Biochem. 2015;116(10):2146–54.
Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282(2):224–58.
Faustova M, Plestilova L, Hulejova H, Pecha O, Betteridge Z, Mann H, Putova I, Vencovsky J, Novota P, Krystufkova O. Genetic variation in promoter sequence of B-cell-activating factor of the TNF family (BAFF) in patients with idiopathic inflammatory myopathies (IIM) http://ard.bmj.com/content/72/Suppl_1/A51.3.full.pdf.
Lin JD, Yang SF, Wang YH, Fang WF, Lin YC, Lin YF, et al. Analysis of associations of human BAFF gene polymorphisms with autoimmune thyroid diseases. PLoS One. 2016;11(5):e0154436.
Boisson B, Wang YD, Bosompem A, Ma CS, Lim A, Kochetkov T, et al. A recurrent dominant negative E47 mutation causes agammaglobulinemia and BCR(−) B cells. J Clin Invest. 2013;123(11):4781–5.
Frasca D, Nguyen D, Riley RL, Blomberg BB. Decreased E12 and/or E47 transcription factor activity in the bone marrow as well as in the spleen of aged mice. J Immunol. 2003;170(2):719–26.
Patel D, Chinaranagari S, Chaudhary J. Basic helix loop helix (bHLH) transcription factor 3 (TCF3, E2A) is regulated by androgens in prostate cancer cells. Am J Cancer Res. 2015;5(11):3407–21.
Kardava L, Yang Q, St Leger A, Foon KA, Lentzsch S, Vallejo AN, et al. The B lineage transcription factor E2A regulates apoptosis in chronic lymphocytic leukemia (CLL) cells. Int Immunol. 2011;23(6):375–84.
Ansell SM, Novak AJ, Ziesmer S, Price-Troska T, LaPlant B, Dillon SR, et al. Serum BLyS levels increase after rituximab as initial therapy in patients with follicular grade 1 non-Hodgkin lymphoma. Am J Hematol. 2009;84(2):71–3.
Eilertsen GO, Van Ghelue M, Strand H, Nossent JC. Increased levels of BAFF in patients with systemic lupus erythematosus are associated with acute-phase reactants, independent of BAFF genetics: a case-control study. Rheumatology. 2011;50(12):2197–205.
Fabris M, Quartuccio L, Vital E, Pontarini E, Salvin S, Fabro C, et al. The TTTT B lymphocyte stimulator promoter haplotype is associated with good response to rituximab therapy in seropositive rheumatoid arthritis resistant to tumor necrosis factor blockers. Arthritis Rheum. 2013;65(1):88–97.
Acknowledgments
This work was supported by the National Science Centre Poland, grant number N N402 680940. Part of patients’ DNA samples and clinical data was obtained in cooperation supported by the National Science Centre Poland, grant number N N402 682040. Intracellular analysis of BAFF was supported by research grant number 2 PO5B 116 27 from the State Committee for Scientific Research (Poland). We wish to thank Izabela Nowak, PhD, and Andrzej Wiśniewski, PhD, from the Laboratory of Immunogenetics and Tissue Immunology for great help in collecting DNA samples from healthy volunteers. We are also very grateful to our patients and healthy volunteers for blood donation and agreement to participate in this study and the following MD doctors for help in collecting patients’ samples and clinical data: Prof. Kazimierz Kuliczkowski; Marek Kiełbiński, MD, PhD (Department of Hematology, Neoplastic Diseases, and Bone Marrow Transplantation, Medical University, ul. Pasteura 1, 50-367 Wroclaw, Poland); Dariusz Woszczyk, MD, PhD (Department of Hematology, State Hospital, ul. Kosnego 53, Opole, Poland); and Prof. Irena Frydecka (Department of Experimental Therapy, Institute of Immunology and Experimental Therapy, Polish Academy of Science, ul. Weigla 12, 53-114 Wroclaw, Poland).
Authors’ contribution
MJ planned the study; ABJ carried out FACS and ELISA experiments; MJ performed the majority of genotyping experiments, data analysis, and interpretation and part of statistical analysis; MW performed part of genotyping experiments and part of statistical analysis; MS performed the majority of statistical analysis; DW and JR were involved in patient recruitment and clinical data collection, data analysis, and revision of the manuscript; MJ wrote the manuscript; MS and ABJ participated in manuscript writing; and LK and PK revised the manuscript. All authors approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Helsinki Declaration and its later amendments or comparable ethical standards.
Electronic supplementary material
ESM 1
(DOCX 102 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jasek, M., Bojarska-Junak, A., Wagner, M. et al. Association of variants in BAFF (rs9514828 and rs1041569) and BAFF-R (rs61756766) genes with the risk of chronic lymphocytic leukemia. Tumor Biol. 37, 13617–13626 (2016). https://doi.org/10.1007/s13277-016-5182-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5182-z