Abstract
Background
Short tandem repeat (STR) markers cannot be used to distinguish between genetically identical monozygotic (MZ) twins, causing problems in a case with an MZ twin as a suspect. Many studies have shown that in older MZ twins, there are significant differences in overall content and genomic distribution of methylation.
Objective
In this study, we analyzed the DNA methylome profile of blood to identify recurrent differentially methylated CpG sites (DMCs) to discriminate between MZ twins.
Methods
Blood samples were collected from 47 paired MZ twins. We performed the DNA methylation profiling using the HumanMethylation EPIC BeadChip platform and identified recurrent DMCs between MZ twins. Then, Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and motif enrichment analyses were performed to reveal the biological functions of recurrent DMCs. We collected DNA methylome data from the Gene Expression Omnibus (GEO) public database to verify the recurrent DMCs between MZ twins.
Results
We identified recurrent DMCs between MZ twin samples and observed that they were enriched in immune-related genes. In addition, we verified our DMCs in a public dataset.
Conclusion
Our results suggest that the methylation level at recurrent DMCs between MZ twins may serve as a valuable biomarker for identification of individuals in a pair of MZ twins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Identifying individuals from a pair of monozygotic (MZ) twins is important in the forensic sciences, particularly for cases concerning an MZ twin as a suspect or alleged parent. Unfortunately, short tandem repeat (STR) markers cannot distinguish MZ twins because they share an identical genomic DNA sequence. Although several studies have reported that MZ twins have a very small degree of genetic differences, such as single-nucleotide polymorphisms (SNPs) (Weber-Lehmann et al. 2014) or copy number variations (CNVs) (Abdellaoui et al. 2015; McRae et al. 2015), these sequence differences are sparse.
DNA methylation, which occurs at the 5’-position of cytosine in CpG dinucleotides, is involved in regulating gene expression (Moore et al. 2013). As individual DNA methylation levels are uniquely changed by several factors such as environment or disease (Fraga et al. 2005; Planterose Jimenez et al. 2021), DNA methylation analysis has emerged as a method for forensic identification of individuals in a paired MZ twins. In an initial study, Xu et al. reported that DNA methylation at LINE-1 has great potential to discriminate between MZ twins, but its difference was observed in only a small percentage of MZ twins (12.61%) (Xu et al. 2015). Leander Stewart et al. proposed DNA methylation markers at the Alu sequences to identify an MZ twin (Stewart et al. 2015).
DNA microarray analysis is a valuable tool for DNA methylome profiling and discovering new biomarkers. Several researchers have used DNA microarrays to identify MZ twins. Du et al. reported 38 differentially methylated regions in four MZ twins by methylated DNA immunoprecipitation (MeDIP) (Du et al. 2015). Furthermore, Vidaki et al. conducted DNA methylome profiling in 10 pairs of female MZ twins using HumanMethylation 450 K BeadChip and reported twin-differentially methylated sites (Vidaki et al. 2017). Currently, many genome-wide DNA methylome profiling techniques are available, and using more samples for DNA methylome profiling is helpful to identify recurrent DNA methylation markers for discrimination of MZ twins.
To identify potential DNA methylation markers to discriminate MZ twins, we performed genome-wide DNA methylome profiling with 47 paired MZ twin samples using Illumina HumanMethylation EPIC BeadChip, which includes over 850,000 CpG sites. We identified the 26,874–147,713 CpG sites from each pair of MZ twins, and then selected 8,583 recurrent differentially methylated CpG sites (DMCs) between MZ twins. Furthermore, our recurrent DMCs between MZ twins were validated in another cohort from GEO. Our data will be helpful for the development of DNA methylation markers for use in forensic science.
Materials and methods
Study subjects and DNA isolation
Blood samples were collected from 94 healthy volunteers in a Korean cohort after obtaining informed consent from the participants. This study was approved by the Institute Review Board of the National Biobank of Korea (https://www.nih.go.kr/biobank/) and Seoul National University Hospital (C-2110-183-1267). Genomic DNA was isolated from blood using DNeasy Blood and Tissue Kit (Qiagen, Carlsbad, CA). The quality and quantity of the extracted genomic DNA were assessed with an ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE).
DNA methylome profiling and data analysis
HumanMethylation EPIC BeadChip (Illumina, San Diego, CA) was used for DNA methylome screening according to the manufacturer’s instructions. For DNA methylome data analysis, CpG methylation values were calculated as average-β values using the minfi package (version 1.26.2) (Aryee et al. 2014) of R software, and we used the subset-quantile within array normalization (SWAN) to reduce technical variations within and between arrays (Maksimovic et al. 2012). Measurements with detection P-values < 0.05 were considered to have a significant signal above the background.
Public data collection
To validate DMCs selected from our cohort, we obtained a DNA methylation dataset of blood from GEO (GSE154566) generated from 118 paired MZ twin samples using the HumanMethylation EPIC BeadChip platform.
Motif and gene enrichment analysis
Analysis of sequence motifs was performed using a HOMER package (version 4.10) with the default parameter settings, using recurrent DMCs (Heinz et al. 2010). Regions for motif analyses were defined as 100 bp upstream to 100 bp downstream of DMCs. We used the EnrichR tool (https://maayanlab.cloud/Enrichr/) (Chen et al. 2013) for Kyoto Encyclopedia of Genes and Genomes (KEGG) Gene ontology (GO) enrichment analysis.
Statistical analysis
R software (version 4.2.2) was used to analyze and plot data. To identify recurrent DMCs between MZ twins, we used standard deviation and methylation differences for each paired MZ twin sample. We employed Student’s t-test to evaluate the difference in the number of DMCs between male and female groups. Pearson’s correlation was performed to confirm the association between age and the number of DMCs. The chi-square test was utilized for the DMCs proportion of CpG density or gene category in genomic regions. Results with P-values < 0.05 were considered significant.
Results
Methylation difference between each paired MZ twin sample
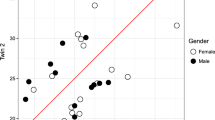
We carried out DNA methylome profiling using the HumanMethylation EPIC BeadChip platform of blood collected from 47 paired MZ twin samples. For identification of DMCs between paired MZ twin samples, we calculated the methylation difference for each paired MZ twin sample. DMCs with methylation difference values higher than 0.05 (average-β values) were considered. As a result, we identified 26,874–147,713 CpG sites from each paired MZ twin sample (Fig. 1a). The number of DMCs was not significantly different between male and female groups (Fig. 1b), but it did increase with age (Fig. 1c).
Identification of recurrent DMCs and their characteristics
For identification of recurrent DMCs between paired MZ twin samples, we applied the following two criteria: (1) DNA methylation standard deviation > 0.05 (average-β values) and (2) average methylation difference > 0.05 (average-β values) between MZ twins. As a result, we identified 8583 recurrent DMCs (Fig. 2a). We analyzed the distribution feature of recurrent DMCs based on the gene structure and CpG island relationship. Gene structures were classified into eight categories: TSS1500, TSS200, 1stExon, body, intergenic, ExonBnd, 3’UTR, and 5’UTR. Our results showed that recurrent DMCs were significantly enriched in the gene body regions compared to the non-DMCs (Fig. 2b). CpG island relationships were classified into four categories: island, shore (up to 2 kb from CpG island), shelf (2–4 kb from CpG island), and open sea (> 4 kb from CpG island). We observed that recurrent DMCs were significantly enriched in open seas compared to non-DMCs (Fig. 2c).
Distribution of recurrent DMCs between MZ twins and their proportion of CpG density of gene category in genomic regions. a Distribution of recurrent DMCs with average methylation differences. b genomic distribution, c CpG relationship. ExonBnd, within 20 bases of an exon boundary; Shore, ~ 0–2 kb from the CGI; shelf, ~ 2–4 kb from the CGI; open sea, > 4 kb from the CGI. CGI, CpG island
Recurrent DMCs associated with the immune system
We performed KEGG pathway, GO, and motif analyses to reveal the biological function of recurrent DMCs in transcription start site (TSS) regions. KEGG pathway analysis revealed allograft rejection, T-cell receptor signaling pathway, asthma, and Th1 and Th2 cell differentiation (Fig. 3a). In GO analysis, the antigen receptor-mediated signaling pathway, T-cell receptor signaling pathway, macrophage activation, and regulation of cytokine production were highlighted (Fig. 3b). Transcription factors are proteins with DNA binding activity that are involved in regulation of transcription. In generally, TFs modulate gene expression by binding to gene promoter regions of distal regions called enhancers. The distance between a TFBS and a TSS of a gene regulated by a TF can be up to several megabases, and depends on the chromatin structures of the regions (Dekker and Heard 2015). To examine whether the recurrent DMCs identified are associated with TFBSs, we performed TF motif analysis using the 8,583 recurrent DMCs. Our results revealed the recurrent DMCs to be enriched in CEBP, NFIL3, HLF, Atf4, and ERG and involved in regulation of the immune system (Kim et al. 2019; Ng et al. 2020; Poli 1998; Yang et al. 2018). The top 10 binding motifs for the recurrent DMCs are shown in Fig. 3c. These results indicated that the recurrent DMCs are related to the immune system.
Enrichment analysis using recurrent DMCs between MZ twins. a GO and b KEGG pathway analyses using the EnrichR tool. For GO and KEGG enrichment analyses, 1210 recurrent DMCs located at TSS were used. c Recurrent DMCs of known transcription factor binding sites based on HOMER packages with default parameter settings. Sequences within + 100 bp or − 100 bp flanking each of the recurrent DMCs were used for known motif analyses
Validation of recurrent DMC in the public dataset
To validate our recurrent DMCs, we collected DNA methylome data from the GEO public database and then analyzed the public methylome data in the same way. Of our 8583 recurrent DMCs, 7149 were validated in the public dataset (Fig. 4a). The methylation difference in each paired MZ twin samples of the top 4 recurrent DMCs (cg12597325, cg01095518, cg17310354, and cg09042759) is shown in Fig. 4b. The top 20 candidate recurrent DNA methylation markers to discriminate between MZ twins are summarized in Table 1. The results suggest that DNA methylation has a great potential application for forensic identification of individuals in an MZ twin pair.
Discussion
Tissue-specific patterns of DNA methylation have been demonstrated (Kitamura et al. 2007; Song et al. 2009). In this study, we chose to base our analysis on blood samples because blood is commonly found at crime scenes. To identify potential DNA methylation markers for identifying an individual in a pair of MZ twins, we conducted genome-wide DNA methylation profiling with 47 paired MZ twin samples using HumanMethylaton EPIC BeadChip. Then, we observed tens to hundreds of thousands of DMCs for each individual paired MZ twin samples. The change in DNA methylation between MZ twins was not significantly different between male and female groups, but it correlated with age. Our result is similar to previous results (Kim et al. 2018; Martin 2005).
The immune system specializes in responding to environmental exposures (MacGillivray and Kollmann 2014), and DNA methylation plays an important role in the immune system by controlling gene expression through changes in chromatin structure(Morales-Nebreda et al. 2019). KEGG and GO analyses showed recurrent DMCs between MZ twins to be enriched in the immune system. Enhancers or superenhancers (Sur and Taipale 2016) are also associated with DNA methylation (Heyn et al. 2016), and our motif analysis showed the recurrent DMCs to be enriched at immune system-associated transcription factor-binding sites such as CEBP (Larabee et al. 2018), NFIL3 (Schlenner et al. 2019), and ATF (Jadhav and Zhang 2017).
Several techniques are available for measuring DNA methylation levels (Laird 2010). In this study, we identified recurrent DMCs, but the difference in methylation level of most recurrent DMCs was less than 0.1 (βvalues). Therefore, it is essential to choose a validation method for DNA methylation with few technical errors. Further study will be needed to validate our recurrent DMCs by validation techniques such as pyrosequencing (Ronaghi 2001) and amplicon bisulfite sequencing (Masser et al. 2013), as these techniques provide high-accuracy methylation measurements and require small amounts of DNA.
In conclusion, we expect that our DNA methylation study and novel DNA methylation markers will help to improve the use of DNA to discriminate individuals in a pair of monozygotic twins.
Data availability
All primary methylation array data were deposited in the GEO (https://www.ncbi.nlm.nih.gov/geo/) public database under Accession Number GSE225544.
References
Abdellaoui A, Ehli EA, Hottenga JJ, Weber Z, Mbarek H, Willemsen G, van Beijsterveldt T, Brooks A, Hudziak JJ, Sullivan PF et al (2015) CNV Concordance in 1097 MZ Twin Pairs. Twin Res Hum Genet 18:1–12
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA (2014) Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30:1363–1369
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128
Dekker J, Heard E (2015) Structural and functional diversity of topologically associating domains. FEBS Lett 589:2877–2884
Du Q, Zhu G, Fu G, Zhang X, Fu L, Li S, Cong B (2015) A genome-wide scan of dna methylation markers for distinguishing monozygotic twins. Twin Res Hum Genet 18:670–679
Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J et al (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 102:10604–10609
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589
Heyn H, Vidal E, Ferreira HJ, Vizoso M, Sayols S, Gomez A, Moran S, Boque-Sastre R, Guil S, Martinez-Cardus A et al (2016) Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol 17:11
Jadhav K, Zhang Y (2017) Activating transcription factor 3 in immune response and metabolic regulation. Liver Res 1:96–102
Kim S, Wyckoff J, Morris AT, Succop A, Avery A, Duncan GE, Jazwinski SM (2018) DNA methylation associated with healthy aging of elderly twins. Geroscience 40:469–484
Kim HS, Sohn H, Jang SW, Lee GR (2019) The transcription factor NFIL3 controls regulatory T-cell function and stability. Exp Mol Med 51:1–15
Kitamura E, Igarashi J, Morohashi A, Hida N, Oinuma T, Nemoto N, Song F, Ghosh S, Held WA, Yoshida-Noro C et al (2007) Analysis of tissue-specific differentially methylated regions (TDMs) in humans. Genomics 89:326–337
Laird PW (2010) Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 11:191–203
Larabee JL, Hauck G, Ballard JD (2018) Unique, intersecting, and overlapping roles of C/EBP beta and CREB in cells of the innate immune system. Sci Rep 8:16931
MacGillivray DM, Kollmann TR (2014) The role of environmental factors in modulating immune responses in early life. Front Immunol 5:434
Maksimovic J, Gordon L, Oshlack A (2012) SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol 13:R44
Martin GM (2005) Epigenetic drift in aging identical twins. Proc Natl Acad Sci U S A 102:10413–10414
Masser DR, Berg AS, Freeman WM (2013) Focused, high accuracy 5-methylcytosine quantitation with base resolution by benchtop next-generation sequencing. Epigenetics Chromatin 6:33
McRae AF, Visscher PM, Montgomery GW, Martin NG (2015) Large autosomal copy-number differences within unselected monozygotic twin pairs are rare. Twin Res Hum Genet 18:13–18
Moore LD, Le T, Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38
Morales-Nebreda L, McLafferty FS, Singer BD (2019) DNA methylation as a transcriptional regulator of the immune system. Transl Res 204:1–18
Ng AP, Coughlan HD, Hediyeh-Zadeh S, Behrens K, Johanson TM, Low MSY, Bell CC, Gilan O, Chan YC, Kueh AJ et al (2020) An Erg-driven transcriptional program controls B cell lymphopoiesis. Nat Commun 11:3013
Planterose Jimenez B, Liu F, Caliebe A, Montiel Gonzalez D, Bell JT, Kayser M, Vidaki A (2021) Equivalent DNA methylation variation between monozygotic co-twins and unrelated individuals reveals universal epigenetic inter-individual dissimilarity. Genome Biol 22:18
Poli V (1998) The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem 273:29279–29282
Ronaghi M (2001) Pyrosequencing sheds light on DNA sequencing. Genome Res 11:3–11
Schlenner S, Pasciuto E, Lagou V, Burton O, Prezzemolo T, Junius S, Roca CP, Seillet C, Louis C, Dooley J et al (2019) NFIL3 mutations alter immune homeostasis and sensitise for arthritis pathology. Ann Rheum Dis 78:342–349
Song F, Mahmood S, Ghosh S, Liang P, Smiraglia DJ, Nagase H, Held WA (2009) Tissue specific differentially methylated regions (TDMR): changes in DNA methylation during development. Genomics 93:130–139
Stewart L, Evans N, Bexon KJ, van der Meer DJ, Williams GA (2015) Differentiating between monozygotic twins through DNA methylation-specific high-resolution melt curve analysis. Anal Biochem 476:36–39
Sur I, Taipale J (2016) The role of enhancers in cancer. Nat Rev Cancer 16:483–493
Vidaki A, Diez Lopez C, Carnero-Montoro E, Ralf A, Ward K, Spector T, Bell JT, Kayser M (2017) Epigenetic discrimination of identical twins from blood under the forensic scenario. Forensic Sci Int Genet 31:67–80
Weber-Lehmann J, Schilling E, Gradl G, Richter DC, Wiehler J, Rolf B (2014) Finding the needle in the haystack: differentiating “identical” twins in paternity testing and forensics by ultra-deep next generation sequencing. Forensic Sci Int Genet 9:42–46
Xu J, Fu G, Yan L, Craig JM, Zhang X, Fu L, Ma C, Li S, Cong B (2015) LINE-1 DNA methylation: a potential forensic marker for discriminating monozygotic twins. Forensic Sci Int Genet 19:136–145
Yang X, Xia R, Yue C, Zhai W, Du W, Yang Q, Cao H, Chen X, Obando D, Zhu Y et al (2018) ATF4 Regulates CD4 (+) T Cell Immune Responses through Metabolic Reprogramming. Cell Rep 23:1754–1766
Acknowledgements
This work was supported by the grants from the National Research Council of Science & Technology (NTC0022211) and Korean National Police Agency (PR10-01-000-21). This study was conducted with bioresources from the National Biobank of Korea, the Korea Disease Control and Prevention Agency, Republic of Korea (NBK-2022-046).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no conflict interests.
Ethical approval
This study was in compliance with the code of ethics for human rights mentioned in the 7th revision of the 1975 Declaration of Helsinki (2013), and the protocols were ratified by the Ethics Committee of National Biobank of Korea. All participants enrolled were informed and provided the signed the consent form.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, JY., Lee, H.Y., Lee, SY. et al. DNA methylome profiling of blood to identify individuals in a pair of monozygotic twins. Genes Genom 45, 1273–1279 (2023). https://doi.org/10.1007/s13258-023-01396-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-023-01396-4