Abstract
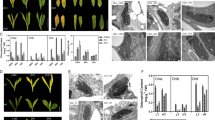
Normal and the spontaneous spirally rolled leaves of Cymbidium goeringii var. longibracteatum were used for RNA sequencing analyses using the Illumina paired-end sequencing technique to figure out the differently-expressed genes in two samples. About 5.65 and 4.82 Gb sequencing data of raw reads were obtained from 2 cDNA libraries of normal and the spirally rolled leaves respectively. After data filtering, quality checks and de novo assembly, a total of 48,935 unigenes with an average sequence length of 820 nt were generated. In addition, the transcriptome change in normal and the spirally rolled leaves was investigated. With non-redundant annotation, 219 differentially expressed genes (DEGs) are identified, with 147 up-regulated genes and 72 down-regulated genes. Out of these DEGs, 21 DEGs (9.59 %) were involved in cell wall modeling enzymes, such as expansin, xyloglucan endo-transglycosylase, pectate lyase, cell wall-associated hydrolase. Besides, other DEGs were predominantly classified as genes involved in transcription factor and signal sense and transduction signaling. This study presents the first comprehensive characterization of the leave transcriptomes of Cymbidium goeringii var. longibracteatum. This study not only gave us valuable sequence resources of this species, but also provided theoretical foundation for cultivar breeding of leaf mutation in C. goeringii var. longibracteatum.




Similar content being viewed by others
References
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188
Berná G, Robles P, Micol JL (1999) A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152:729–742
Chen H, Zha J, Liang X, Bu J, Wang M, Wang Z (2013) Sequencing and de novo assembly of the Asian clam (Corbicula fluminea) transcriptome using the Illumina GAIIx method. PLoS ONE 8:e79516
Chen Y, Mao Y, Liu H, Yu F, Li S, Yin T (2014) Transcriptome analysis of differentially expressed genes relevant to variegation in peach flowers. PLoS One 9:e90842
Conesa A, Götz S, García-Gómez J, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Cosgrove DJ (2000) Expansive growth of plant cell walls. Plant Physiol Biochem 38:109–124
Cosgrove DJ, Durachko DM (1994) Autolysis and extension of isolated walls from growing cucumber hypocotyls. J Exp Bot 45:1711–1719
Cosgrove DJ, Li LC, Cho H-T, Hoffmann-Benning S, Moore RC, Blecker D (2002) The growing world of expansins. Plant Cell Physiol 43:1436–1444
Grabherr MG, Haas BJ, Moran Y, Levin JZ, Thompson DA, Ido A, Xian A, Lin F, Raktima R, Qiandong Z (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Hew CS (2001) Ancient Chinese orchid cultivation: a fresh look at an age-old practice. Sci Hortic 87:1–10
Ichihashi Y, Horiguchi G, Gleissberg S, Tsukaya H (2010) The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol 51:252–261
Jones AM, Im KH, Savka MA, Wu MJ, Dewitt NG, Shillito R, Binns AN (1998) Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282:1114–1117
Kim HJ, Lee JS, Park KW, Kim SS (2013) New cultivar ‘White Edge’ of leaf variegated Hosta minor. Korean J Plant Resources 26:516–518
Lee Y, Choi D, Kende H (2001) Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 4:527–532
Li Y, Jones L, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6:603–610
Lomax J (2005) Get ready to GO! A biologist's guide to the gene ontology. Brief Bioinform 6:298–304
Luo Z, Yang Z, Zhong B, Li Y, Xie R, Zhao F, Ling Y, He G (2007) Genetic analysis and fine mapping of a dynamic rolled leaf gene, RL10(t), in rice (Oryza sativa L.). Genome 50:811–817
Mcqueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425–1433
Mcqueen-Mason SJ, Fry SC, Durachko DM, Cosgrove DJ (1993) The relationship between xyloglucan endotransglycosylase and in vitro cell wall extension in cucumber hypocotyls. Planta 190:327–331
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Nishihara M, Yamada E, Saito M, Fujita K, Takahashi H, Nakatsuka T (2014) Molecular characterization of mutations in white-flowered torenia plants. BMC Plant Biol 14:1–13
Pérez-Pérez JM, Ponce MR, Micol JL (2001) ULTRACURVATA1, a SHAGGY-like Arabidopsis gene required for cell elongation. Int J Dev Biol 45:S51–S52
Pérez-Pérez JM, MaR Ponce, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242:161–173
Qiu WM, Zhu AD, Yao W, Chai LJ, Ge XX, Deng XX, Guo WW (2012) Comparative transcript profiling of gene expression between seedless Ponkan mandarin and its seedy wild type during floral organ development by suppression subtractive hybridization and cDNA microarray. BMC Genom 13:157
Robles P, Micol J (2001) Genome-wide linkage analysis of Arabidopsis genes required for leaf development. Mol Genet Genomics 266:12–19
Sara Z, Alberto F, Enrico G, Luciano X, Marianna F, Giovanni M, Diana B, Mario P, Massimo D (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152:1787–1795
Schunmann PHD, Smith RC, Lang V, Matthews PR, Chandler PM (1997) Expression of XET-related genes and its relation to elongation in leaves of barley (Hordeum vulgare L.). Plant, Cell Environ 20:1439–1450
Shao YJ, Chen ZX, Zhang YF, Chen EH, Ding Cheng QI, Miao J, Pan XB (2005) One major QTL mapping and physical map construction for rolled leaf in rice. Acta Genet Sin 32:501–506 (in Chinese with English Abstract)
Singh JP, Arora RS, Dohare SR, Sengupta K (1970) A spontaneous mutant for flower colour and shape in a white flowering dahlia. Euphytica 19:261–262
Tasaki K, Nakatsuka A, Cheon KS, Kobayashi N (2015) Inheritance of the narrow leaf mutation in traditional Japanese evergreen azaleas. Euphytica 206:649–656
Wang B, Li Z, Yu C (2005) Progress on orchid breeding Study. Acta Hort Sin 32:551–556
Wang HZ, Wu ZX, Lu JJ, Shi NN, Zhao Y, Zhang ZT, Liu JJ (2009) Molecular diversity and relationships among Cymbidium goeringii cultivars based on inter-simple sequence repeat (ISSR) markers. Genetica 136:391–399
Xie F, Burklew C, Yang Y, Liu M, Xiao P, Zhang B, Qiu D (2012) De novo sequencing and a comprehensive analysis of purple sweet potato (Impomoea batatas L.) transcriptome. Planta 236:101–113
Yang T, Poovaiah B (2000) Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem 275:3137
Yang C, Li D, Liu X, Ji C, Hao L, Zhao X, Li X, Chen C, Cheng Z, Zhu L (2014) OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol 14:158
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Youn-Sung K, Kim S-G, Park J-E, Park H-Y (2006) A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18:3132–3144
Zhang C, Wang Y, Fu J, Dong L, Gao S, Du D (2014a) Transcriptomic analysis of cut tree peony with glucose supply using the RNA-Seq technique. Plant Cell Rep 33:111–129
Zhang YJ, Wang XJ, Wu JX, Chen SY, Chen H, Chai LJ, Yi HL (2014b) Comparative transcriptome analyses between a spontaneous late-ripening sweet orange mutant and its wild type suggest the functions of ABA, sucrose and JA during citrus fruit ripening. PLoS ONE 9:e116056
Zhang C, Wang Y, Fu J, Bao Z, Zhao H (2016) Transcriptomic analysis and carotenogenic gene expression related to petal coloration in Osmanthus fragrans ‘Yanhong Gui’. Trees. doi:10.1007/s00468-016-1359-8
Zhao D, Jiang Y, Ning C, Meng J, Lin S, Ding W, Tao J (2014) Transcriptome sequencing of a chimaera reveals coordinated expression of anthocyanin biosynthetic genes mediating yellow formation in herbaceous peony (Paeonia lactiflora Pall.). BMC Genom 15:689
Zhou H-G, Wang W-T, Qiao Z-Q, Wang Y-J, Wang H-C, Wang F-L, Huang Z-F, He Z-M (2015) A new chrysanthemum cultivar ‘Binfen’. Acta Hort Sin 42:201–202 (in Chinese with English Abstract)
Acknowledgments
This work was supported by the Flower Breeding of New Varieties Project for Zhejiang Science and Technology Department (Grant No. 2009C12087).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Huijuan Ning declares that she has no conflict of interest, Chao Zhang declares that he has no conflict of interest, Jianxin Fu declares that she has no conflict of interest, Yirong Fan declares that he has no conflict of interest.
Ethical Approval
The article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ning, H., Zhang, C., Fu, J. et al. Comparative transcriptome analysis of differentially expressed genes between the curly and normal leaves of Cymbidium goeringii var. longibracteatum . Genes Genom 38, 985–998 (2016). https://doi.org/10.1007/s13258-016-0443-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-016-0443-1