Abstract
Spatially explicit estimates of population density, together with appropriate estimates of uncertainty, are required in many management contexts. Density surface models (DSMs) are a two-stage approach for estimating spatially varying density from distance sampling data. First, detection probabilities—perhaps depending on covariates—are estimated based on details of individual encounters; next, local densities are estimated using a GAM, by fitting local encounter rates to location and/or spatially varying covariates while allowing for the estimated detectabilities. One criticism of DSMs has been that uncertainty from the two stages is not usually propagated correctly into the final variance estimates. We show how to reformulate a DSM so that the uncertainty in detection probability from the distance sampling stage (regardless of its complexity) is captured as an extra random effect in the GAM stage. In effect, we refit an approximation to the detection function model at the same time as fitting the spatial model. This allows straightforward computation of the overall variance via exactly the same software already needed to fit the GAM. A further extension allows for spatial variation in group size, which can be an important covariate for detectability as well as directly affecting abundance. We illustrate these models using point transect survey data of Island Scrub-Jays on Santa Cruz Island, CA, and harbour porpoise from the SCANS-II line transect survey of European waters. Supplementary materials accompanying this paper appear on-line.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Distance sampling is a widely used method for estimating abundance when detection is imperfect (Buckland et al. 2001), based on encounters along line or point transects. Detection probability (detectability) is estimated using within-encounter data (e.g. perpendicular distance from trackline), by fitting “detection functions” that may involve environmental covariates (e.g. local weather conditions). In traditional stratified distance sampling, an average animal density is then estimated within each survey stratum—i.e. some region within which survey coverage is supposed to be uniform—based on the observed encounter rate within that stratum divided by the detectability, and then scaled by the stratum area. Since the abundance estimate is a simple function of statistically independent quantities (encounter rate and detectability), its variance can be estimated straightforwardly.
Instead of using strata, with modern statistical tools it is possible to fit spatially explicit models of density, where local density is assumed to vary gradually in space (and perhaps also in response to specific environmental covariates, which we here include under the general heading of “spatially explicit”). Spatially explicit estimates are advantageous in many situations: when abundance estimates are required across arbitrary sub-regions that do not coincide with survey strata; to reduce bias when coverage is uneven; or when identifying particularly important habitat for conservation, for example.
There are various approaches to actually fitting spatially explicit models. The general idea, as in the stratified case, is that the expected local encounter rate is the product of local detectability and local density, but with both factors now potentially depending on local spatial and/or environmental covariates. Here, we consider specifically density surface models (DSMs; Hedley and Buckland 2004; Miller et al. 2013), which take a two-stage approach. The first stage is to estimate detectability using a detection function model; any standard or bespoke model could be used (see Sect. 2). In the second stage, the encounter rate data are fitted to location/environmental covariates using a GAM, specifically the “basis-and-penalty” formulation of GAMs in Wood (2017) in which smoothers are represented via random effects. The estimated detectabilities for each segment of search effort are easily accommodated in the GAM (technically, as offsets to the linear predictor; see below), and the range of smoothers and interactions that can be fitted in is very wide.
Splitting the analysis into two stages is appealing partly because existing domain-specific software and diagnostic expertise can be applied as-is to each stage separately, and partly because it avoids any need to write inevitably complicated code that incorporates two individually complex aspects. It is also straightforward to produce a point estimate of abundance for any desired sub-region straight from the fitted GAM. However, when detectability and density both vary spatially, the problem is what to do about variance given that GAMs do not intrinsically “understand” the notion of uncertainty in their offset.
In this paper, we show how statistical uncertainty about detectability can in fact be accommodated painlessly within standard GAM software. Our approach is to first fit the detection function as usual, but then to rewrite the fitted detection function log-likelihood as a quadratic approximation centred on its point estimates, and to incorporate the uncertainty about the detection function parameters via random effects in the second-stage GAM. This fits directly and automatically into the Wood/Wahba formulation of a GAM, whereby a smooth surface is described by a set of coefficients treated as random effects; thus, the machinery for handling random effects in general is already built into the mgcv software used in Miller et al. (2013)’s DSM code. This amounts to refitting the detection function model (or a good approximation to it) at the same time as fitting the GAM. We retain the benefits of two-stage modelling, but all the uncertainty about detectability as well as density is now captured in the usual GAM outputs. The refitted detection function model should not differ greatly from the original fit, and we use this idea to propose a diagnostic for overall model specification issues.
Following a summary of DSMs and notation in Sect. 2, we present our new formulation in Sect. 3, including variance computation and diagnostics. Section 4 comments on problems with existing approaches to variance propagation in DSMs. In Sect. 5, we extend the formulation to cover DSMs where group size varies spatially and affects detectability (a common situation with whales and dolphins). Section 6 gives examples of the variance propagation method and the group size model. Some discussion is given in Sect. 7, including possible generalizations.
2 Density Surface Models
In distance sampling, observers move along a set of survey lines or between points, counting (groups of) animals, recording distances from the centre line or centre point to the observed groups (or their cues, such as blows for cetaceans or calls for birds), the size of each detected group and potentially other covariates that may affect detectability.
To fully describe the DSMs in this paper, we distinguish four different classes of variable.
-
1.
Density covariates, x, vary in space and potentially affect local animal abundance: e.g. latitude and depth. They are required for prediction and fitting, and are assumed known across the entire region of interest.
-
2.
Effort covariate(s), z, affect detection probability: e.g. sea conditions measured on the Beaufort scale, or observer identity. They are assumed known along each transect, but not necessarily in unsurveyed areas.
-
3.
Individual covariates, g, that affect detection probability and are a persistent property of each group (independent of whether the group is observed or not) during its window of observability: e.g. size (number of animals), and perhaps behaviour. Here, g is assumed known for each observed group (see Discussion). The random variable G varies from one group to the next, and its statistical distribution \(F_{G}\left( g;x\right) \) may vary spatially. \(F_{G}\left( g;x\right) \) may have a direct effect on abundance (via the mean group size), as well as on detection probability, in which case it is also necessary to estimate certain properties of \(F_{G}\left( g;x\right) \) such as its local mean.
-
4.
Observation variables, y, which are random properties of one observation on one group: e.g. perpendicular distance between the group and the sampler. In certain settings, y may contain other elements. For example, in a multi-observer platform survey (e.g. MRDS; Borchers et al. 1998), y might also include which of the active observers saw the group; in a cue-based setting, y might include the bearing between sighting and observer.
These classes are assumed to be mutually exclusive; overlaps can lead to fundamental problems for distance sampling which we do not address here (e.g. non-uniform animal distribution within the sample unit; Marques et al. 2012). The distinction between individual and effort covariates is often glossed over but they have rather different implications for abundance estimation (see below).
In the first stage of DSM, the detection function \(\pi \left( y|\theta ,z,g\right) \), which involves unknown parameters \(\theta \) as well as z and g, describes the probability of making an observation at y. The parameters \(\theta \) are usually estimated by maximizing this log-likelihood across observations s:
where \(t_{s}\) is the transect containing sighting s. Here, p is the overall detection probability for a group, defined by
where \(F_{Y}\) is the distribution function of y. In standard distance sampling where y consists only of perpendicular distance, \(F_{Y}\) is uniform between 0 and some fixed truncation distance, beyond which observations are discarded. This formulation encompasses a wide range of models, including multiple covariate distance sampling (MCDS; Marques and Buckland 2003) with z and g, multi-observer mark–recapture distance sampling (MRDS; Borchers et al. 1998), and cue-based “hazard probability models” (Skaug and Schweder 1999).
The second part of DSM models the local count of observations via a GAM to capture spatial variation in animal density. This allows us both to estimate abundance within any sub-region of interest, and to compensate as far as possible for uneven survey coverage (whether by design, or by virtue of field logistics and weather conditions). Since line transects are generally very long in comparison with their width and therefore contain a range of density and density covariate values, we divide transects into smaller segments, which are the sample units for GAM (in which case the subscript \(t_{s}\) above refers to segments rather than transects). Point transects are left as-is; we use the term “segments” from now on to refer to both points and line segments, without loss of generality. Environmental covariates are assumed not to change much within each segment. The relationship between counts \(n_{i}\) per segment i and density covariates \(x_{ik}\) is modelled as an additive combination of smooth functions with a \(\log \) link:
where each segment is of area \(a_{i}\), and \(n_{i}\) follows some count distribution such as quasi-Poisson, Tweedie, or negative binomial. The \(f_{k}\) are smooth functions, represented by a basis expansion (\(f_{k}(x)=\sum _{j}\beta _{j}b_{j}(x)\), for some basis functions \(b_{j}\)); \(\beta _{0}\) is an intercept term, included in parameter vector \(\varvec{\beta }\); \(\varvec{\lambda }\) is a vector of smoothing (hyper)parameters which control the wiggliness of the \(f_{k}\). We take a Bayesian interpretation of GAMs, in which \(\varvec{\lambda }\) controls the variance of a multivariate improper Gaussian prior (Wood 2017):
with scale parameter \(\phi \) , smoothing parameters \(\lambda _{k}\) and penalty matrices \(\mathbf {S}_{k}\) (\(^{-}\) indicates pseudoinverse). This leads to a quadratic penalty on beta during fitting. We estimate \(\varvec{\lambda }\) itself via REML (Wood 2011), an empirical Bayes procedure. Fully Bayesian approaches, placing hyperpriors on \(\varvec{\lambda }\) are also possible.
We are interested in the uncertainty of a predicted abundance estimate, \({\hat{N}}\). We assume below that we have created some prediction grid with all density covariates available for each cell in the grid. Abundance is predicted for each cell, and summed for an overall abundance, \({\hat{N}}\), over some region of interest which may not be the entire surveyed area. Although \(p(\hat{\varvec{\theta }})\) does not appear explicitly in the prediction, which is
the GAM offsets \(p(\hat{\varvec{\theta }})\) clearly do affect \(\hat{\varvec{\beta }}\), so it is important to account somehow for detection probability uncertainty. (2) assumes the offset is fixed, so extra steps are required.
3 Variance Propagation for Density Surface Models
Let \(p\left( \varvec{\theta }_{0},z_{i}\right) \) be the true probability of detection in segment i and for now omit g, thereby assuming that there are no individual-level covariates (e.g. that group size is always 1) for now (see Sect. 5). If \(\varvec{\theta }_{0}\) is the true (unknown) value of \(\varvec{\theta }\), and \(\hat{\varvec{\theta }}\) is its MLE, we use the shorthand \(p_{i}=p\left( \varvec{\theta }_{0},z_{i}\right) \) and \({\hat{p}}_{i}=p\left( \hat{\varvec{\theta }},z_{i}\right) \) when the dependence is clear. The expected number of encounters in segment i is \(a_{i}p_{i}\rho _{i}\) where \(\rho _{i}\) is the underlying density, given by the exponential term in 2.
Given \(p_{i}\), we can rewrite (2) on the \(\log \) link scale as:
\(X_{i}\) is the (known) ith row of the design matrix, i.e. the values of the basis functions in segment i, so \(\log \rho _{i}=\sum _{k}f_{k}(x_{ik})=X_{i}\varvec{\beta }\) and \(\log a_{i}p_{i}\) is an offset. The complication is that we only have an estimate of \(p_{i}\). To tackle this, we first rewrite the linear predictor \(\eta _{i}\) as
and then take a Taylor series expansion of \(\log {\hat{p}}_{i}\equiv \log p\left( \hat{\varvec{\theta }},z_{i}\right) \) about \(\varvec{\theta }=\varvec{\theta _{0}}\):
By defining the vectors \(\varvec{\delta }\triangleq \hat{\varvec{\theta }}-\varvec{\theta }_{0}\) and \(\kappa _{i}\triangleq \left. \frac{d\log p\left( \varvec{\theta },z_{i}\right) }{d\varvec{\theta }}\right| _{\varvec{\theta }=\varvec{\theta }_{0}}\), we can rewrite (3) as
In Supplementary Materials C, we show that the approximations in our approach do not affect the asymptotic order of accuracy. Specifically, the Laplace approximation that underlies REML estimation is accurate to \(O\left( n^{-1}\right) \) and our approximations are of the same order (see Supplementary Materials C for the meaning of n).
We have approximately that \(\varvec{\theta }_{0}\vert \mathbf {y}\sim N\left( \hat{\varvec{\theta }},\mathbf {V}_{\varvec{\theta }}\right) \implies \varvec{\delta }\sim N\left( \mathbf {0},\mathbf {V}_{\varvec{\theta }}\right) ,\) where the covariance matrix \(\mathbf {V}_{\varvec{\theta }}\) is calculated as the negative inverse Hessian of (1). In other words, the “posterior distribution” of \(\varvec{\theta }\) from fitting the detection function now becomes a prior distribution for \(\rho \). To first order, \(\varvec{\delta }\) then plays the same structural role in (5) as the basis coefficients \(\varvec{\beta }\). The design matrix for \(\varvec{\delta }\) (\(\kappa \) in (5)) is obtained by differentiating the log-detection probabilities, with respect to \(\varvec{\theta }\) at \(\hat{\varvec{\theta }}\). Simple 3-point numerical differentiation is perfectly adequate for calculation of the derivatives. \(\mathbf {V}_{\varvec{\theta }}\) should be readily available from detection function fitting (via the Hessian) regardless of the complexity of the model.
This method can be applied automatically to almost any distance sampling set-up provided one can calculate detection probabilities, find their derivatives, and obtain a Hessian for the likelihood. Simultaneous estimates \(\varvec{\beta }\) and \(\varvec{\delta }\) can be obtained from standard GAM fitting software. Posterior inferences about \(\varvec{\beta }\) (therefore \(\rho \) and abundance) automatically propagate the uncertainty from fitting the detection function.
The only technical difference from fitting a standard GAM is that \(\varvec{\lambda }\) is usually unknown and has to be estimated (i.e. the prior on \(\varvec{\beta }\) has known covariance, but unknown scale), whereas the prior on \(\varvec{\delta }\) is completely determined from the detection function fitting (i.e. in effect \(\lambda _{\varvec{\delta }}=1/\phi \), where \(\phi \) is the scale parameter). This set-up cannot be specified directly in the R package mgcv because of implementation details (at least up to version 1.8; it may be possible within other GAM implementations), unless \(\phi \) is fixed rather than estimated. This is fine for Poisson or negative binomial response, but in our experience, better fits can often be obtained using a Tweedie response distribution, for which \(\phi \) must be estimated. In order to implement (5) for a general response distribution using mgcv, we therefore use a one-dimensional search over \(\phi \) to maximize the marginal REML. At each iteration, given the working value \(\phi ^{*}\), we refit the GAM fixing \(\phi =\phi ^{*}\) and \(\lambda _{\varvec{\delta }}=1/\phi ^{*}\). Speed can be improved by reusing some of the set-up computations (design matrices, etc.) at each iteration.
Diagnostics. If the detection function fits properly and the spatial model has adequate flexibility, then the second-stage model should not lead to much change in the detection function parameters, so that \(\hat{\varvec{\delta }}\) should be “close” to 0. Nevertheless, there is scope for interaction if the detection function includes covariates that also vary systematically over space. For example, if weather is systematically worse in some parts of the survey region, then both \(\varvec{\beta }\) and \(\varvec{\theta }\) will contribute to the expected pattern of sightings, and the two sets of parameters are partially confounded. (That is of course also true for all-in-one models, as well for our two-stage model.)
There are several diagnostics that we have found useful for checking consistency between the two parts of the model. The first is to compare the inferred spatial distribution and abundance from fitting (3) with the “naïve” estimates where detection uncertainty is ignored and the offset \(a_{i}{\hat{p}}_{i}\) is treated is exact, ensuring that there are not large differences in the estimated spatial distribution. The second is to check whether the detection probabilities (by covariate level) would be substantially changed by fitting the spatial model; in other words, whether \(\hat{\varvec{\delta }}\) is close enough to zero given its prior distribution, or, perhaps more usefully, whether the overall detectability by covariate level has changed. Since the fitted spatial model still includes the information from the first stage, any shift of more than about 1 standard deviations (based on the covariance from the detection function stage) might merit investigation. Third, as a general diagnostic tool for density surface models, we have found it useful to compare total observed and expected numbers of sightings, grouped by detection covariates (e.g. Beaufort). This can be helpful in diagnosing detection function problems, e.g. failure of assumed certain detectability at zero distance under poor weather conditions, as well as failures of the spatial model (e.g. an abrupt change in density). In addition, one could also use standard detection function model checking (e.g. quantile–quantile plots) with the adjusted parameters, \(\hat{\varvec{\theta }}+\hat{\varvec{\delta }}\).
Calculating \(\text {Var}({\hat{N}})\). Once detection function uncertainty has been propagated, we only need to deal with uncertainty in the GAM, which now has an updated covariance matrix. We therefore can rely on two commonly used methods to obtain the variance of model outputs like abundance, \({\hat{N}}\).
1. Delta method We can calculate:
(the delta method) where \(\mathbf {V}_{\hat{\varvec{\beta }}}\) is the covariance matrix for the GAM coefficients Wood (2017, Sects. 5.8 and 6.9.3). We form the prediction matrix, \(\mathbf {X}_{p}\), which maps model coefficients to values of the linear predictor for the prediction data, so \(\varvec{{\hat{\eta }}}_{p}=\mathbf {X}_{p}\varvec{{\hat{\beta }}}\) (Wood 2017, Sect. 6.10). Derivatives are evaluated at the estimated values of the model parameters.
2. Posterior simulation The posterior for \(\varvec{\beta }\) given data \(\mathbf {y}\) and smoothing parameters \(\varvec{\lambda }\) are approximately distributed as \(\varvec{\beta }\vert \mathbf {y},\varvec{\lambda }\sim N(\hat{\varvec{\beta }},\mathbf {V}_{\hat{\varvec{\beta }}})\). The following algorithm then can be used:
-
1.
For \(b=1,\ldots ,B\):
-
(a)
Simulate from \(N(\hat{\varvec{\beta }},\mathbf {V}_{\hat{\varvec{\beta }}})\), to obtain \(\hat{\varvec{\beta }_{b}}\).
-
(b)
Calculate predicted abundance, \({\hat{N}}_{b}=\mathbf {a}_{p}\exp (\mathbf {X}_{p}\hat{\varvec{\beta }_{b}})\) (where \(\mathbf {a}_{p}\) is a row vector of areas for the prediction cells).
-
(a)
-
2.
Calculate the empirical variance or percentiles of the \({\hat{N}}_{b}\)s.
In practice, B in the order of 1000s appears to work well, though there may be some issues when the approximation breaks down. In these cases, we recommend the use of importance sampling (either using importance weights to calculate weighted summaries or using a second resampling of the \({\hat{N}}_{b}\)s) or a Metropolis–Hastings sampler (as implemented in mgcv::gam.mh). Further examples are given in Supplementary Materials.
Software. The procedure given in this section is implemented in the R package dsm, available on CRAN. The dsm_varprop function in the package allows the user to provide a fitted DSM and a prediction grid. Using the delta method, it will then calculate an uncertainty estimate for the estimated abundance for that prediction grid. The function also returns the refitted GAM so one can extract the full covariance matrix and perform posterior simulation if required. Diagnostics for \(\hat{\varvec{\delta }}\) are calculated by a summary method for the returned object.
4 Previous Methods for Estimating Uncertainty in Density Surface Models
Several approaches have previously been suggested to combine detection function and spatial model predicted abundance uncertainties; we review them briefly here. We need to estimate the following:
where P here is a random variable for the (uncertain) probability of detection and the subscripts indicate the expectation/variance taken over that variable. \({\hat{N}}(\{{\hat{p}}_{i};i=1,\ldots ,n\})\) is the estimated abundance as a function of estimated detection probabilities. The first part of this can be derived from GAM theory as shown in the previous section; the second is more tricky.
Assuming independence. When \({\hat{p}}_{i}\) is the same for all observations, then \(N({\hat{p}})\propto 1/{\hat{p}}\), so \({\hat{N}}\) and \({\hat{p}}\) are independent. The total variance of the abundance estimate can be calculated by combining the GAM variance estimate with the variance of the probability of detection summing the squared coefficients of variation (\(\text {CV}(X)=\sqrt{\text {Var}(X)}/{\bar{X}}\)) (Goodman 1960). Hence,
When there are not covariates in the detection function, we calculate:
This is fine when the detection function does not contain any covariates, as there is no covariance then between the effort and density covariates. (The procedure outlined in Sect. 3 does not yield a different answer.) In the case where detectability is a function of covariates, it is impossible in general to justify the use of the CV decomposition as there are correlations between the spatial distribution and the covariates affect detectability.
The approach taken by Program Distance (Thomas et al. 2010) is to use Horvitz–Thompson-adjusted counts per segment, instead of the observed count, as the response in the GAM. Thus removes the detectability from the right hand side of (2). Variance is then calculated by taking the probability of detection averaged over the observations by first calculating the Horvitz–Thompson estimate of the abundance in the covered area (\({\hat{N}}=\sum _{i}g_{i}/\hat{p_{i}}\), where \(g_{i}\) is group size of the \(i^{\text {th}}\) observation and \(\hat{p_{i}}\) is the probability of detecting that group) then using that \({\hat{N}}\) to calculate the implied average detectability, had the analysis not contained covariates (\({\tilde{p}}={\tilde{n}}/{\hat{N}}\), where \({\tilde{n}}\) is the number of observed groups). The numerical derivatives of \({\tilde{p}}\) with respect to \(\varvec{\theta }\) can then be used in (7) to derive a variance for this probability of detection, averaged over the observations.
We do not recommend this approach either. Transforming the response through multiplication by a random variable breaks the mean–variance and independence assumptions of the GAM, so that the computed \(\text {CV}\left( {\hat{N}}_{\text {GAM}}\right) \) is invalid when detection covariates are present. Additionally, there is no coherent way to generalize the formula to small-area predictions—the effort covariates within a small area will not have the same range as those in the larger survey area (e.g. weather conditions will not be homogenous throughout the survey area). Hence, the uncertainty that applies to the overall \({\tilde{p}}\) is usually not the appropriate uncertainty to apply to a small area where observing conditions may be atypical.
The bootstrap. Bootstraps are sometimes seen as an attractive alternative to deal with all aspects of variance in DSMs. Hedley and Buckland (2004) describe two possible implementations (one parametric, one nonparametric), which are not easy to choose between and which do not necessarily give similar answers. Ignoring computational time issues, the first practical difficulty in setting up a “good” nonparametric bootstrap for a DSM is sampling units, independent of the fitted model.
The second, more substantial, issue is the fundamental statistical problem with combining smoothers with bootstraps. The problem does not seem to be well known in the statistical ecology literature, so we give an explanation here. The basic problem is that (most) bootstraps use only the posterior modes of random effects (smooths), thus omitting a key part of the posterior uncertainty. To see this, consider a simple “spatial model” where the region is divided into blocks, each with its own independent random effect, and a bootstrap that generates new data at each original observation/transect, either parametrically or nonparametrically. If one of the blocks is unsampled in the original data, it will be unsampled in every realization too, and the “spatial model” simply sets the point estimate of that random effect to zero in every bootstrap realization; hence, a bootstrap will ascribe zero uncertainty for the density in that block. The correct inference would of course be for the random effect to retain its prior variance.
This phenomenon has been well known in statistics since at least Laird and Louis (1987) (see also the discussants), who coined the term “naïve bootstrap” for such procedures that ignore the point estimate shrinkage inevitable in mixed or random effect models. (Fixed effect models are not susceptible in the same way.) They proposed some parametric modifications (“type II” and “type III” bootstraps) that are more effective in the IID and block-structured situations that they consider. However, the underlying theory is complex (Carlin and Gelfand 1991; Carlin and Louis 2008) and it is far from clear whether simple yet reliable bootstraps can be devised for complicated multi-stage random effect situations like DSMs. Figure 1 shows a simple unidimensional Poisson process, sampled at either end but not in the middle (rug plot). Bootstrap replicates (shown in light grey, of which there are 500) largely fail to capture our uncertainty in the unsampled middle area. The analytical estimate (dark grey band) illustrates how little we know about the unsampled area.
Comparison of bootstrap and analytical uncertainty for a Poisson process. The black line is the true intensity function (on the response scale) and points are observations. Blue line is a smooth of space, light grey wiggly lines are 500 bootstrap predictions, dashed lines are point-wise upper and lower 95% quantiles from the bootstrap, and the dark grey band is the analytical GAM confidence band using (6). The bootstrap appears confident that there is nothing in the unsampled area, but the analytical estimate illustrates how little we know (Color figure online)
The above does not imply that simple or indeed complicated bootstraps will never give reliable results in DSMs; given plenty of observations and good, uniform coverage, many approaches to inference will give similar and good results. However, it is sometimes not obvious whether this holds for a specific dataset, nor what to do bootstrap-wise if not. Instead, the (empirical) Bayesian framework of GAMs offers a coherent and general-purpose way to capture uncertainty.
5 A New Model for Group Size
Our variance propagation method so far works if detectability depends only on effort covariates, but not for individual covariates such as group size. Incorporating individual covariates in the detection function is not problematic, but it is not obvious how to allow for these different detection probabilities in the GAM. Further, it is not obvious how to combine predictions of different group sizes since average group sizes may vary spatially.
One approach is to use the Horvitz–Thompson-adjusted response described in the previous section, but as mentioned above this does not allow variance propagation. One could fit separate spatial models to subsets of the data for each group size, but it seems inefficient to not share information between subsets of the data. Next, we show instead how to extend our variance propagation method to deal with group size.
We form M categories of group sizes, denoted \(\{g_{m};m=1,\ldots ,M\}\), where groups within each category have similar detectability, and fit a detection function incorporating these group size categories. We then fit a GAM to an M-fold replicate of the dataset, with the response in the \(m^{\text {th}}\) replicate of the \(i^{\text {th}}\) segment being \(n_{im}\), the number of groups in category m that were seen in that segment. (The total number of observations is unchanged; each observation is allocated to just one of the “replicates”.) Group size category (as a factor) is included as an explanatory variable, and smooths are modified to allow similar variations in density of groups with different sizes. There are no extra assumptions in this formulation from the model in Sect. 3, except to assume that the numbers of groups of different size categories in a given segment are independent, given the underlying density (which is allowed to vary with group size).
Factor-smooth interactions. We extend (2) to include multiple smooths of space which correspond to different categorizations of group size, so our model is:
for \(m=1,\ldots ,M\) where \(n_{i,g_{m}}\) is the number of observed groups in group class \(g_{m}\) in segment i and \(f_{x_{1},g_{m}}\) is the spatial smooth (where \(x_{1}\) is a spatial coordinate) for group size class \(g_{m}\). Smoothers like \(f_{x_{1},g_{m}}\) are referred to as factor-smooth interactions (Wood 2017; Pedersen et al. 2019). \(f_{k}\) are any other smooths (of covariates \(x_{k}\), for \(k>1\)). For clarity, we make the dependence on group size class explicit: \(p({\hat{\theta }};z_{i},g_{m})\), i.e. the probability of detection given segment-level detection covariates \(z_{i}\) and group size \(g_{m}\).
There are a number of different possible forms for \(f_{x_{1},g_{m}}\). These vary in two main ways: (1) do levels share a smoothing parameter, or have separate ones? (2) do smooths tend towards a “global” smooth that dictates a general spatial effect? Here, we adopt the “fs” basis in mgcv which can be thought of as a smooth version of a random slopes model: smooths are generated for each factor level with smooths defined as deviations from a reference level, with all smooths sharing the same smoothing parameter. This is appealing as we might expect that the spatial smooths for each group size are similar but there might be some process that generates larger groups in certain places (e.g. large prey aggregations attracting large groups of animals). This approach is easily extended to other density covariates (e.g. \(x_{1}\) could be bathymetry or vegetation cover).
Abundance and uncertainty estimation with group size smooths. Abundance is estimated by summing over the predictions for each group size category (\({\hat{N}}_{m}\)) and weighting them by the corresponding mean group size (\({\bar{g}}_{m}\)): \({\hat{N}}=\sum _{m=1}^{M}{\bar{g}}_{m}{\hat{N}}_{m}.\) We can find \(\text {Var}({\hat{N}}\vert \bar{G)}\) (where \({\bar{G}}\) is the mean group size) from the variance propagation procedure above, but we need \(\text {Var}({\hat{N}})\), which we can obtain from the law of total variance:
where \(\mathrm {Var}({\bar{G}}_{m})\) reflects the uncertainty about mean group size within a category, to be estimated empirically from all the observed groups in that category. The effect of \(\mathrm {Var}({\bar{G}}_{m})\) on \(\mathrm {Var}(N)\) should be small (because categories are narrow, and mean must lie within category), and also should not vary much spatially, so no further spatial adjustment to that variance component is required.
6 Examples
Island Scrub-Jays. We first apply our variance propagation method in a simple situation where there is covariance between the abundance and detection processes that is the case of a spatially varying detection covariate. Island Scrub-Jays (Aphelocoma insularis) are endemic to Santa Cruz Island, California. Jays primarily reside in areas of chaparral and forest, though the density of this foliage also affects detectability. Sillett et al. (2012) model the distribution Island Scrub-Jays from 307 point transects surveyed in fall 2008 and spring 2009. Distances were binned into three intervals due to responsive movement (\([0m-100m],(100m-200m],(200m-300m]\)). Proportion chaparral (\(\texttt {chap}\)) and proportion forest (\(\texttt {forest}\)) were available as covariates, as was elevation (\(\texttt {elev}\)). Sillett et al. fitted a hierarchical model assuming a negative binomial distribution for abundance and a multi-nomial detection process using a half-normal detection function. Their best models (by AIC) were: fall 2008 abundance modelled as \(\beta _{0}+\beta _{1}{} \texttt {\texttt {chap}}^{2}+\beta _{2}{} \texttt {chap}+\beta _{3}{} \texttt {elev}\), with detectability as a function of \(\texttt {chap}\); spring 2009 abundance modelled as \(\beta _{0}+\beta _{1}{} \texttt {chap}^{2}+\beta _{2}{} \texttt {chap}+\beta _{3}{} \texttt {elev}^{2}+\beta _{4}{} \texttt {elev}\), detectability as a function of \(\texttt {forest}\).
We replicated the analysis of Sillett et al. using our two-stage variance propagation approach to show that our method can be used in such a situation. In summary, final coefficient estimates were very close to those in the original paper, abundance estimates with associated 95% CIs were very similar for both seasons: Fall 2008 DSM \({\hat{N}}=\)2272 (1625–3175), Sillet et al. \({\hat{N}}=\)2267 (1613–3007) and Spring 2009 DSM \({\hat{N}}=\)1684 (1263-2246), Sillet et al. \({\hat{N}}=\)1705 (1212–2369); Supplementary Material A gives the comparison in full. For the spring model, the value of the \(\texttt {forest}\) coefficient in the detection function changed effect size from \(-0.18\) (SE = 0.06) to \(-0.083\) (SE = 0.062) after propagation (indicating no issue with our \(\hat{\varvec{\delta }}\) diagnostic). By giving the GAM the flexibility to slightly adjust the detection function parameters via \(\hat{\varvec{\delta }}\) (as opposed to treating the estimated detection probabilities as certain), the CV of the abundance estimate is actually improved in this case, from 18.4 to 14.8%.
The jay data present a particularly interesting case as the covariates in the GAM are fixed effects; there is therefore no “cost” (in terms of the penalized likelihood) to changing the GAM coefficients. We see minimal changes in the parameters of the fall model (Supplementary Material A, Table 1), as these are already well modelled (no doubt due to the good coverage of the data): the detection function includes \(\texttt {chap}\) and the GAM includes \(\texttt {chap}\) and \(\texttt {chap}^{2}\), so any adjustment via \(\hat{\varvec{\delta }}\) is a third-order effect. The spring model has different covariates in each model component, making the correction necessary.
The survey design had extremely good coverage over Santa Cruz Island. We decided to see what the effect of “unbalancing” the design would be to test the robustness of our model. We randomly sub-sampled the fall data to contain only 100 sites and then removed those where chaparral cover was greater than the mean chaparral proportion (over all points). Our sub-sample was left with 16 detections at 65 points. Fitting the fall DSM to the reduced data yields \({\hat{N}}=\)26,434 (95% CI 209-3,349,000; CV=2,100%). Post-variance propagation, we obtain \({\hat{N}}=\)2,831 (95% CI 39-206,000; CV=1,100%), both detection function and GAM coefficients having changed (see Supplementary Material A, Table 3). While we would expect a high variance for such a small and unbalanced dataset (and indeed we obtain this), our procedure tames the model to an extent, giving a more realistic estimate of abundance. Once information about both model components is allowed to inform the parameter estimation simultaneously, the coefficients are corrected.
Island Scrub-Jays Simulation. To assess performance of our variance propagation method with the delta method and a one-stage fully Bayesian approach, we conducted a simulation using the Island Scrub-Jay data as a starting point. We kept spatial coverage constant throughout the simulation settings but varied the detectability and therefore the number of observations available for the detection function component of the model. Full details of the simulation set-up are given in Supplementary Material B. Here, we note that our variance propagation method performed well in terms of bias in the abundance estimate and its corresponding variance estimate compared to the fully Bayesian model, even when sample size decreased (Supplementary Material B, Figure 2).
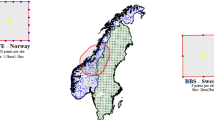
Harbour porpoise. To illustrate our new group size model, we reanalyse an aerial line transect survey of harbour porpoise in Irish Sea, coastal Irish waters, and Western coastal Scotland, where we see spatial variation in observed group size of 1 to 5 animals (typical for harbour porpoise; e.g. Siebert et al. 2006; points in Fig. 2). During SCANS-II aerial surveys, two observers recorded cetacean detections (along with sighting conditions) from bubble windows on both sides of a plane flying at 183m. Complete survey details and a comprehensive analysis is given in Hammond et al. (2013). For simplicity we assume certain detection on the trackline, no errors in group size estimation (less likely with aerial than in shipboard surveys for harbour porpoise; Phil Hammond, Debi Palka, pers. comm., November 2017), and negligible island/coastline effects in the spatial model.
To fit our DSM, three group size bins were formed: size 1 (131 observations), 2 (35 observations) and 3-5 (14 observations). A hazard rate detection function was fitted to the observed distances (truncated at 300m) with the group size bin (\(g_{m}\), \(m=1,\ldots ,3\)) and Beaufort (\(B_{i}\), binned as 0-1, 2 and 3-5) as factor covariates. Detectability for each segment i per group size factor was then estimated from the detection function: \(p(\hat{\varvec{\theta }};B_{i},g_{m})\). Following (8), we fitted the DSM:
where in segment i of area \(a_{i}\) the observed number of groups in size category \(g_{m}\) was denoted \(n_{i,g_{m}}\). It was assumed the response was Tweedie-distributed where the power parameter was constrained to be greater than 1.2 to avoid numerical issues. Each \(f_{E,N,g_{m}}\) was a smooth of space (projected Easting/Northing; \(E_{i}\), \(N_{i}\)) for group category \(g_{m}\) and had a maximum basis size of 20 (total maximum basis size for \(f_{E,N,g_{m}}\) was therefore 60).
Predicted density surfaces from the new group size model for harbour porpoise. First three plots are density maps for the given group size (i.e. group abundance multiplied by mean group size), right plot shows the combined map, summing the previous three plots per prediction cell. We can see that distribution is roughly similar in all three group size categories though with almost no larger groups in the North, far more animals occurring as singletons than in larger groups
The fitted model had a total effective degrees of freedom of 20.47 for \(f_{E,N,g_{m}}\). GAM checking showed reasonable fit to the data. Table 1 shows observed vs expected counts by Beaufort—there is some misfit at the highest state, perhaps because detection probability at zero distance changes with Beaufort level (from Hammond et al.: 0.45 for Beaufort 0-1 and 0.31 in Beaufort 2-3). Plots of the per-group size bin predictions and the combined prediction are given in Fig. 2, which show some consistent patterns between group size classes (“hotspot” off Southern Ireland) and some differences (varying distribution in the Irish Sea and Western Scotland), this kind of insight is not possible using a single smooth for all observations and may prove useful in cases where there are occasional very large group sizes (e.g. oceanic dolphins). Using (7), the CV of abundance was estimated to be 2.36%, when our new variance propagation method was used the CV was estimated as 9.65%. The assumption of independence (via (7)) underestimates uncertainty in the case where group size (and detectability) vary in space. Only a small piece of code implementing (9) was required in addition to the dsm_varprop function (included in Supplementary Material A), we then gain the ability to make inferences about group size spatial distribution (traditionally requiring two separate models, one for encounter rate, one for group size; e.g. Becker et al. 2014), as well as improve uncertainty estimation via variance propagation.
7 Discussion
Combining the uncertainty from detection functions with that from spatial models has been a challenging problem for point and line transect analysis, requiring either complicated bespoke software that combines two model components, or ad hoc approaches that lack statistical justification. In this paper, we have demonstrated a simple, flexible, and statistically sound method that can (i) propagate uncertainty from detectability models to the spatial models for a particular class of detection function (i.e. those without individual-level covariates) and (ii) include group size as a covariate in the detection function while still being able to propagate uncertainty and address spatial variation in group size. Our methods are implemented in the dsm package for R but can be implemented in any standard GAM fitting software.
It is straightforward to apply our factor-smooth approach group size more generally to individual-level covariates which affect detectability and vary in space, but do not directly affect abundance, such as observable behaviour. For example, feeding groups might be more (or less) conspicuous than resting groups, and the proportion feeding/resting may vary across the surveyed region. Unless detectability is included in the analysis, biased abundance estimates could result, especially when survey coverage is non-uniform; and there has been no simple way until now to include such effects in the spatial model. A major advantage of our approach over simple (or complex) stratification schemes is that we are now sharing information between the levels of our categorized variable. This makes the results less sensitive to over-specifying the number of categories, as the model will shrink back towards the simpler model in the absence of strongly informative data. We also note that the factor-smooth approach could be applied to all smooth terms in the GAM, allowing for a very flexible model. This would be appropriate only if it was reasonable that all smooths vary according to the detectability covariate (e.g. feeding behaviour in our harbour porpoise example might depend on both space and depth).
We have assumed that all variables are measured without much error. Measurement error for individual-level covariates such as group size can be a serious problem in distance sampling (Hodgson et al. 2017)—distance between observer and group can affect not just detectability, but also the extent of group size error. If group size varies spatially, it is hard to see how to separate the spatial modelling stage from the distance sampling stage. A full discussion is beyond the scope of this paper, but we suspect that specially designed observation protocols and bespoke analyses may be the only way to tackle such thorny cases.
All-in-one fitting of both detection and spatial models is also possible (e.g. Johnson et al. 2009; Sillett et al. 2012; Yuan et al. 2017). If models are specified correctly, then the all-in-one approach could in theory be slightly more efficient, but only insofar as it takes account of third-order changes in the detection function likelihood (since our approach uses a quadratic approximation). That seems unlikely to make much difference in general—and as is the case for the Island Scrub-jay example. Our own preference is therefore to use the two-stage approach, mainly because in our experience the careful fitting of detection functions is a complicated business which can require substantial model exploration and as few as possible “distractions” (such as simultaneously worrying about the spatial model). The two-stage process allows any form of detection function to be used, without having to make deep modifications to software. In summary, if one knew one had the correct model to begin with, one-stage fitting would be slightly more efficient, but this is never the case in practice.
It is valuable to check for any tension or confounding between the detection function and density surface parts of the model, which can occur if there are large-scale variations in sighting conditions across the survey region, and which is readily diagnosed in a two-stage model. Although this does not appear to lead to problems in the datasets we have analysed with the software described in this paper, we have come across it in other variants of line transect-based spatial models with different datasets. It may not be so easy to detect partial confounding when using all-in-one frameworks.
Finally, we note that the approach outlined here (of using a first-stage estimate as a prior for a second estimate, and propagating variance appropriately) is quite general and is comparable to standard sequential Bayesian approaches to the so-called integrated data models. The first-stage model need not be a detection function, but instead could be from another GAM (or other latent Gaussian model). Again, this allows us to ensure that first-stage models are correct before moving to more complex modelling. Modelling need not only be of two stages and could extend to multi-stage models (Hooten et al. 2019).
Change history
03 April 2021
A Correction to this paper has been published: https://doi.org/10.1007/s13253-021-00449-z
References
Becker EA, Forney KA, Foley DG, Smith RC, Moore TJ, Barlow J (2014) Predicting seasonal density patterns of California cetaceans based on habitat models. Endanger Species Res 23(1):1–22
Borchers DL, Zucchini W, Fewster RM (1998) Mark-recapture models for line transect surveys. Biometrics 54(4):1207
Buckland ST, Anderson DR, Burnham KP, Borchers DL, Thomas L (2001) Introduction to distance sampling. Estimating abundance of biological populations. Oxford University Press, Oxford
Carlin BP, Gelfand AE (1991) A sample reuse method for accurate parametric empirical Bayes confidence intervals. J R Stat Soc Ser B
Carlin BP, Louis TA (2008) Bayesian methods for data analysis, 3rd edn. CRC Press, Boca Raton
Goodman LA (1960) On the exact variance of products. J Am Stat Assoc 55(292):708
Hammond PS, Macleod K, Berggren P, Borchers DL, Burt L, Cañadas A et al (2013) Cetacean abundance and distribution in European Atlantic shelf waters to inform conservation and management. Biol Conserv 164(C):107–122
Hedley SL, Buckland ST (2004) Spatial models for line transect sampling. J Agric Biol Environ Stat 9(2):181–199
Hodgson A, Peel D, Kelly N (2017) Unmanned aerial vehicles for surveying marine fauna: assessing detection probability. Ecol Appl Publ Ecol Soc Am 27(4):1253–1267
Hooten MB, Johnson DS, Brost BM (2019) Making recursive Bayesian inference accessible. The American Statistician
Johnson DS, Laake JL, Ver Hoef JM (2009) A model-based approach for making ecological inference from distance sampling data. Biometrics 66(1):310–318
Laird NM, Louis TA (1987) Empirical Bayes Confidence Intervals Based on Bootstrap Samples. J Am Stat Assoc 82(399):739–750
Marques FFC, Buckland ST (2003) Incorporating covariates into standard line transect analyses. Biometrics 59(4):924–935
Marques TA, Buckland ST, Bispo R, Howland B (2012) Accounting for animal density gradients using independent information in distance sampling surveys. Stat Methods Appl 22(1):67–80
Miller DL, Burt ML, Rexstad EA, Thomas L (2013) Spatial models for distance sampling data: recent developments and future directions. Methods Ecol Evol 4(11):1001–1010
Pedersen EJ, Miller DL, Simpson GL, Ross N (2019) Hierarchical generalized additive models: an introduction with mgcv. PeerJ, p e6876
Siebert U, Gilles A, Lucke K, Ludwig M, Benke H, Kock K-H, Scheidat M (2006) A decade of harbour porpoise occurrence in german waters-analyses of aerial surveys, incidental sightings and strandings. J Sea Res 56(1):65–80
Sillett TS, Chandler RB, Royle JA, Kéry M, Morrison SA (2012) Hierarchical distance-sampling models to estimate population size and habitat-specific abundance of an island endemic. Ecol Appl 22(7):1997–2006
Skaug HJ, Schweder T (1999) Hazard models for line transect surveys with independent observers. Biometrics 55(1):29–36
Thomas L, Buckland ST, Rexstad EA, Laake JL, Strindberg S, Hedley SL et al (2010) Distance software: design and analysis of distance sampling surveys for estimating population size. J Appl Ecol 47(1):5–14
Wood SN (2011) Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J R Stat Soc Ser B Stat Methodol 73(1):3–36
Wood SN, (2017) Generalized Additive Models: An Introduction with R, Second Edition. Chapman and Hall/CRC Texts in Statistical Science. CRC Press, Boca Raton
Yuan Y, Bachl FE, Lindgren F, Borchers DL, B IJ, Buckland ST et al (2017) Point process models for spatio-temporal distance sampling data from a large-scale survey of blue whales. Ann Appl Stat 11(4):2270–2297
Acknowledgements
The authors thank Natalie Kelly, Jason Roberts, Eric Rexstad, Phil Hammond, Steve Buckland, and Len Thomas for useful discussions, Devin Johnson for the suggestion of this as a general statistical method, and Simon Wood for continued development of mgcv and GAM theory. The manuscript was great improved by comments from the editor and two anonymous reviewers. Data from the SCANS-II project were supported by the EU LIFE Nature programme (project LIFE04NAT/GB/000245) and governments of range states: Belgium, Denmark, France, Germany, Ireland, Netherlands, Norway, Poland, Portugal, Spain, Sweden, and UK. This work was funded by OPNAV N45 and the SURTASS LFA Settlement Agreement, and being managed by the US Navy’s Living Marine Resources program under Contract No. N39430-17-C-1982, US Navy, Chief of Naval Operations (Code N45), grant number N00244-10-1-0057 and the International Whaling Commission.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
M. V. Bravington and D. L. Miller: Joint first author.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bravington, M.V., Miller, D.L. & Hedley, S.L. Variance Propagation for Density Surface Models. JABES 26, 306–323 (2021). https://doi.org/10.1007/s13253-021-00438-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13253-021-00438-2