Abstract
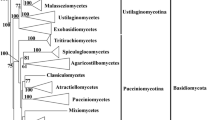
Zygomycetes are phylogenetically early diverging, ecologically diverse, industrially valuable, agriculturally beneficial, and clinically pathogenic fungi. Although new phyla and subphyla have been constantly established to accommodate specific members and a subkingdom Mucoromyceta, comprising Calcarisporiellomycota, Glomeromycota, Mortierellomycota and Mucoromycota, was erected to unite core zygomycetous fungi, phylogenetic relationships within phyla have not been well resolved. Taking account of the information of monophyly and divergence time estimated from ITS and LSU rDNA sequences, the present study updates the classification framework of the phylum Mucoromycota from the class down to the generic rank: three classes, three orders, 20 families (including five new families Circinellaceae, Protomycocladaceae, Rhizomucoraceae, Syzygitaceae and Thermomucoraceae) and 64 genera. The taxonomic hierarchy was calibrated with estimated divergence times: phylum earlier than 617 Mya, classes and orders earlier than 547 Mya, families earlier than 199 Mya, and genera earlier than 12 Mya. Along with this outline, all genera of Mucoromycota are annotated and 58 new species are described. In addition, three new combinations are proposed. In this study, we update the taxonomic backbone of the phylum Mucoromycota and reinforce its phylogeny. We also contribute numerous new taxa and enrich the diversity of Mucoromycota.









































































Similar content being viewed by others
Data availability
All sequences have been deposited at GenBank database (Table 1 and S1).
References
Abrashev R, Krumova E, Petrova P et al (2021) Distribution of a novel enzyme of sialidase family among native filamentous fungi. Fungal Biol 125:412–425. https://doi.org/10.1016/j.funbio.2020.12.006
Agrawal S, Nandeibam J, Devi I (2021) Danger of exposure to keratinophilic fungi and other dermatophytes in recreational place in the northeast region of India. Aerobiologia 37:755–766. https://doi.org/10.1007/S10453-021-09719-2
Agrios GN (2005) Plant pathology, 5th edn. Academic Press, San Diego
Alastruey-Izquierdo A, Hoffmann K, de Hoog GS et al (2010) Species recognition and clinical relevance of the zygomycetous genus Lichtheimia (syn. Absidia pro parte, Mycocladus). J Clin Microbiol 48:2154–2170. https://doi.org/10.1128/JCM.01744-09
Alpat S, Alpat SK, Çadırcı BH et al (2008) A novel microbial biosensor based on Circinella sp. modified carbon paste electrode and its voltammetric application. Sens Actuat B 134:175–181. https://doi.org/10.1016/j.snb.2008.04.044
Alvarez E, Cano J, Stchigel AM et al (2011) Two new species of Mucor from clinical samples. Med Mycol 49:62–72. https://doi.org/10.3109/13693786.2010.499521
Alvarez E, Stchigel AM, Cano J et al (2010) Molecular phylogenetic diversity of the emerging mucoralean fungus Apophysomyces: proposal of three new species. Rev Iberoam Micol 27:80–89. https://doi.org/10.1016/j.riam.2010.01.006
Amos RE, Barnett HL (1966) Umbelopsis versiformis, a new genus and species of the imperfects. Mycologia 58:805–808. https://doi.org/10.2307/3756856
André LDA, Hoffmann K, Lima DX et al (2014) A new species of Lichtheimia (Mucoromycotina, Mucorales) isolated from Brazilian soil. Mycol Prog 13:343–352. https://doi.org/10.1007/s11557-013-0920-8
Babu AG, Kim SW, Adhikari M et al (2015) A new record of Gongronella butleri isolated in Korea. Mycobiology 43:166–169. https://doi.org/10.5941/MYCO.2015.43.2.166
Bai FR, Yao S, Cai CS et al (2021) Mucor rongii sp. nov., a new cold-tolerant species from China. Curr Microbiol 78:2464–2469. https://doi.org/10.1007/s00284-021-02494-w
Baijal U, Mehrotra B (1965) Species of Mucor from India 2. Sydowia 19:204–212
Bainier G (1903) Sur quelques especes de Mucorinées nouvelles ou peu connues. Bull Soc Mycol France 19:153–172
Barraclough T, Nee S (2001) Phylogenetics and speciation. Trends Ecol Evol 16:391–399. https://doi.org/10.1016/S0169-5347(01)02161-9
Batra LR, Segal RH, Baxter RW (1964) A new Middle Pennsylvanian fossil fungus. Am J Bot 51:991–995. https://doi.org/10.1002/j.1537-2197.1964.tb06728.x
Baxter RW (1975) Fossil fungi from America Pennsylvanian coal balls. The University of Kansas Paleontological Contributions 7:1–6
Beauverie J (1900) Mycocladus Verticillatus Ann Univ Lyon 3:162–180
Benjamin CR, Hesseltine CW (1957) The genus Actinomucor. Mycologia 49:240–249. https://doi.org/10.1080/00275514.1957.12024635
Benjamin CR, Hesseltine CW (1959) Studies on the genus Phycomyces. Mycologia 5:751–771. https://doi.org/10.1080/00275514.1959.12024858
Benjamin RK (1959) The merosporangiferous mucorales. Aliso 4:321–433
Benjamin RK (1978) Gamsiella, a new subgenus of Mortierella (Mucorales: Mortierellaceae). Aliso 9:157–170
Benny GL (1991) Gilbertellaceae, a new family of the Mucorales (Zygomycetes). Mycologia 83:150–157. https://doi.org/10.1080/00275514.1991.12025991
Benny GL (1992) Observations on Thamnidiaceae (Mucorales) 5. Thamnidium Mycologia 84:834–842. https://doi.org/10.1080/00275514.1992.12026214
Benny GL (1995a) Kirkomyces, a new name for Kirkia Benny. Mycologia 87:922. https://doi.org/10.1080/00275514.1995.12026615
Benny GL (1995b) Observations on Thamnidiaceae 7. Helicostylum and a new genus Kirkia. Mycologia 87:253–264. https://doi.org/10.1080/00275514.1995.12026527
Benny GL, Benjamin RK (1975) Observations on Thamnidiaceae (Mucorales). New taxa, new combinations, and notes on selected species. Aliso 8:301–351
Benny GL, Benjamin RK (1976) Observations on Thamnidiaceae (Mucorales) 2. Chaetocladium, Cokeromyces, Mycotypha, and Phascolomyces. Aliso 8:391–424
Benny GL, Benjamin RK (1991) The Radiomycetaceae (Mucorales; Zygomycetes) 3. A new species of Radiomyces, and cladistic analysis and taxonomy of the family; with a discussion of evolutionary ordinal relationships in Zygomycotina. Mycologia 83:713–735. https://doi.org/10.2307/3760429
Benny GL, Benjamin RK (1993) Observations on Thamnidiaceae (Mucorales) 6. Two new species of Dichotomocladium and the zygospores of D. hesseltinei (Chaetocladiaceae). Mycologia 85:660–671. https://doi.org/10.2307/3760511
Benny GL, Blackwell M (2004) Lobosporangium, a new name for Echinosporangium Malloch, and Gamsiella, a new genus for Mortierella multidivaricata. Mycologia 96:143–149. https://doi.org/10.1080/15572536.2005.11833004
Benny GL, Humber RA, Morton JB (2001) Zygomycota: zygomycetes. In: McLaughlin DJ, McLaughlin EG, Lemke PA (eds) Systematics and evolution, 1st edn. Springer, Berlin Heidelberg, Berlin, Heidelberg, pp 113–146
Benny GL, Humber RA, Voigt K (2014) Zygomycetous fungi: phylum entomophthoromycota and subphyla kickxellomycotina, mortierellomycotina, mucoromycotina, and zoopagomycotina. In: Mclaughlin DJ, Spatafora JW (eds) Systematics and evolution, 2nd edn. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 209–250. https://doi.org/10.1007/978-3-642-55318-9_8
Benny GL, Kirk PM, Samson RA (1985) Observations on Thamnidiaceae (Mucorales) 3. Mycotyphaceae fam. nov. and a re-evaluation of Mycotypha sensu Benny & Benjamin illustrated by two new species. Mycotaxon 22:119–148
Benny GL, Schipper M (1992) Observations on Thamnidiaceae (Mucorales) 4. Pirella Mycologia 84:52–63. https://doi.org/10.1080/00275514.1992.12026103
Berbee ML, James TY, Strullu-Derrien C (2017) Early diverging fungi: diversity and impact at the dawn of terrestrial life. Ann Rev Microbiol 71:41–60. https://doi.org/10.1146/annurev-micro-030117-020324
Berbee ML, Taylor JW (2001) Fungal molecular evolution: gene trees and geologic time. In: McLaughlin DJ, McLaughlin EG, Lemke PA (eds) Systematics and evolution, 1st edn. Springer, Berlin, pp 229–245
Bergman K, Burke PV, Cerdá-Olmedo E et al (1969) Phycomyces. Bacteriol Rev 33:99–157
Berney C, Pawlowski J (2006) A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc R Soc B 273:1867–1872. https://doi.org/10.1098/rspb.2006.3537
Blair J (2009) Fungi. In: Hedges SB, Kumar K (eds) The timetree of life, 1st edn. Oxford University Press, New York, pp 215–129
Boedijn KB (1958) Notes of the Mucorales of Indonesia. Sydowia 12:321–362
Bonifaz A, Stchigel AM, Guarro J et al (2014) Primary cutaneous mucormycosis produced by the new species Apophysomyces mexicanus. J Med Mycol 52:4428–4431. https://doi.org/10.1128/JCM.02138-14
Bouckaert R, Heled J, Kühnert D et al (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computati Biol 10:e1003537. https://doi.org/10.1371/journal.pcbi.1003537
Brooks FT, Hansford CG (1923) Mould growths upon cold-store meat. Trans Br Mycol Soc 8:113–142. https://doi.org/10.1016/S0007-1536(23)80020-1
Chakrabarti A, Shivaprakash MR, Curfs-Breuker I et al (2010) Apophysomyces elegans: epidemiology, amplified fragment length polymorphism typing, and in vitro antifungal susceptibility pattern. J Med Mycol 48:4580–4585. https://doi.org/10.1128/JCM.01420-10
Chander J, Singla N, Kaur M et al (2021) Apophysomyces variabilis, an emerging and worrisome cause of primary cutaneous necrotizing infections in India. J Mycol Med 31:101197. https://doi.org/10.1016/J.MYCMED.2021.101197
Chang Y, Desirò A, Na H et al (2019) Phylogenomics of Endogonaceae and evolution of mycorrhizas within Mucoromycota. New Phytol 222:511–525. https://doi.org/10.1111/nph.15613
Chang Y, Wang S, Sekimoto S et al (2015) Phylogenomic analyses indicate that early fungi evolved digesting cell walls of algal ancestors of land plants. Genome Biol Evol 7:1590–1601. https://doi.org/10.1093/gbe/evv090
Chen FJ (1992a) Haplosporangium—a new record genus of Mucorales from China. Mycosystema 5:19–22
Chen FJ (1992b) Mortierella species in China. Mycosystema 5:23–64
Cheng Y, Gao Y, Liu XY et al (2017) Rhinocerebral mucormycosis caused by Rhizopus arrhizus var. tonkinensis. J Mycol Med 27:586–588. https://doi.org/10.1016/j.mycmed.2017.10.001
Chibucos MC, Soliman S, Gebremariam T et al (2016) An integrated genomic and transcriptomic survey of mucormycosis-causing fungi. Nat Commun 7:1–11. https://doi.org/10.1038/ncomms12218
Chien CY, Hwang BC (1997) First record of the occurrence of Sporodiniella umbellata (Mucorales) in Taiwan. Mycoscience 38:343–346. https://doi.org/10.1007/BF02464094
Chitnis MV, Munro CA, Brown AJP et al (2002) The zygomycetous fungus, Benjaminiella poitrasii contains a large family of differentially regulated chitin synthase genes. Fungal Genet Biol 36:215–223. https://doi.org/10.1016/S1087-1845(02)00015-4
Coemans E (1863) Quelques hyphomycetes nouveaux 1. Mortierella polycephala et Martensella pectinata. Bulletin de l’Académie Royale des Sciences de Belgique Classe des Sciences 2, ser 15:536–544.
Cooper JA, Park D (2020) Modicella albostipitata, a new species of sporocarp-forming fungus from New Zealand (Mortierellaceae: Mortierellomycota). Phytotaxa 453:293–296. https://doi.org/10.11646/phytotaxa.453.3.11
Corda A (1842) Incones fungorum hucusque cognitorum. Vol. 5. F. Ehrlich, Prague. pp 92.
Cordeiro TRL, Nguyen TTT, Lima DX et al (2020) Two new species of the industrially relevant genus Absidia (Mucorales) from soil of the Brazilian Atlantic Forest. Acta Bot Bras 34:549–558. https://doi.org/10.1590/0102-33062020abb0040
Corrochano LM, Kuo A, Marcet-Houben M et al (2016) Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr Biol 26:1577–1584. https://doi.org/10.1016/j.cub.2016.04.038
Crous PW, Cowan D, Maggs-Kolling G et al (2020) Fungal planet description sheets: 1112–1181. Persoonia 45:251–409. https://doi.org/10.3767/PERSOONIA.2020.45.10
Crous PW, Wingfield MJ, Burgess TI et al (2017) Fungal planet description sheets: 625–715. Persoonia 39:270–467. https://doi.org/10.3767/persoonia.2017.39.11
Crous PW, Wingfield MJ, Burgess TI et al (2018) Fungal planet description sheets: 716–784. Persoonia 40:240–393. https://doi.org/10.3767/persoonia.2018.40.10
Cruz-Lachica I, Marquez-Zequera I, Garcia-Estrada RS et al (2015) First report of Gilbertella persicaria causing papaya fruit rot. Plant Dis 100:227–227. https://doi.org/10.1094/pdis-05-15-0607-pdn
Dai YC, Cui BK, Si J et al (2015) Dynamics of the worldwide number of fungi with emphasis on fungal diversity in China. Mycol Prog 14:1–9. https://doi.org/10.1007/s11557-015-1084-5
Dai YC, Yang ZL, Cui BK et al. (2021) Diversity and systematics of the important macrofungi in Chinese forests. Mycosystema 40:770–805. https://doi.org/10.13346/j.mycosystema.210036
Darriba D, Posada D, Kozlov AM et al (2020) ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol 37:291–294. https://doi.org/10.1093/molbev/msz189
Dashdorj D, Tripathi VK, Cho S et al (2016) Dry aging of beef: review. J Anim Sci Technolol 58:20. https://doi.org/10.1186/s40781-016-0109-1
Davel G, Featherston P, Fernández A et al (2001) Maxillary sinusitis caused by Actinomucor elegans. J Clin Microbiol 39:740–742. https://doi.org/10.1128/JCM.39.2.740-742.2001
Davies JL, Ngeleka M, Wobeser GA (2010) Systemic infection with Mortierella wolfii following abortion in a cow. Can Vet J 51:1391–1393
Davoust N, Persson A (1992) Effects of growth morphology and time of harvesting on the chitosan yield of Absidia repens. Appl Microbiol Biot 37:572–575. https://doi.org/10.1007/BF00240727
de Carvalho Tavares IM, Umsza-Guez MA, Martin N et al (2020) The improvement of grape juice quality using Thermomucor indicae-seudaticae pectinase. J Food Sci Tech 57:1565–1573. https://doi.org/10.1007/s13197-019-04192-9
de Lima CL, Lima DX, de Souza CA et al (2018) Description of Mucor pernambucoensis (Mucorales, mucoromycota), a new species isolated from the Brazilian upland rainforest. Phytotaxa 350:274. https://doi.org/10.11646/phytotaxa.350.3.6
de Lima CLF, Lundgren JDAL, Nguyen TTT et al (2022) Two new species of Backusella (Mucorales, Mucoromycota) from soil in an upland forest in Northeastern Brazil with an identification key of Backusella from the Americas. J Fungi 8:1038. https://doi.org/10.3390/jof8101038
de Souza JI, Marano AV, Pires-Zottarelli CLA et al (2014) A new species of Backusella (Mucorales) from a Cerrado reserve in Southeast Brazil. Mycol Prog 13:981. https://doi.org/10.1007/s11557-014-0981-3
de Souza JI, Pires-Zottarelli CL, Dos Santos JF et al (2012) Isomucor (Mucoromycotina): a new genus from a Cerrado reserve in state of Sao Paulo, Brazil. Mycologia 104:232–241. https://doi.org/10.3852/11-133
Desirò A, Rimington WR, Jacob A et al (2017) Multigene phylogeny of Endogonales, an early diverging lineage offungi associated with plants. IMA Fungus 8:245–257. https://doi.org/10.5598/imafungus.2017.08.02.03
Dixon-Stewart D (1932) Species of Mortierella isolated from soil. Trans Br Mycol Soc 17:208–220
Doilom M, Guo JW, Phookamsak R et al (2020) Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front Microbiol 11:585215. https://doi.org/10.3389/fmicb.2020.585215
Domsch KH, Gams W, Anderson TH (1980) Compendium of soil fungi, vol 1. Academic Press, London. https://doi.org/10.1016/0016-7061(82)90042-8
Ekpo E, Young T (1979) Fine-structure of the dormant and germinating sporangiospore of Syzygites megalocarpus (Mucorales) with notes on Sporodiniella umbellata. Microbios Letters 10:63–68
Ellenberger S, Burmester A, Wöstemeyer J (2016) Complete mitochondrial DNA sequence of the mucoralean fungus Absidia glauca, a model for studying host-parasite interactions. Genome Announc 4:e00153-e216. https://doi.org/10.1128/genomeA.00153-16
Ellis JJ, Hesseltine CW (1965) The genus Absidia: globose-spored species. Mycologia 57:222–235. https://doi.org/10.1080/00275514.1965.12018205
Ellis JJ, Hesseltine CW (1966) Species of Absidia wirh ovoid sporangiospores 2. Sabouraudia 5:59–77. https://doi.org/10.1080/00362176785190311
Ellis JJ, Hesseltine CW (1969) Two new members of the Mucorales. Mycologia 61:863–872. https://doi.org/10.1080/00275514.1969.12018810
Ellis JJ, Hesseltine CW (1974) Two new families of Mucorales. Mycologia 66:87–95. https://doi.org/10.1080/00275514.1974.12019576
Ellis M (1940) Some fungi isolated from pinewood soil. Trans Br Mycol Soc 24:87–97. https://doi.org/10.1016/S0007-1536(40)80022-3
Embree RW (1959) Radiomyces, a new genus in the Mucorales. Am J Bot 46:25–30. https://doi.org/10.1002/j.1537-2197.1959.tb06977.x
Embree RW, Indoh H (1967) Aquamortierella, a new genus in the Mucorales. Bull Torrey Bot Club 94:464–467. https://doi.org/10.2307/2483563
Evans EH (1971) Studies on Mortierella ramanniana 1. Relationship between morphology and cultural behaviour of certain isolates. Trans Br Mycol Soc 56:201–213. https://doi.org/10.1016/S0007-1536(71)80031-1
Gade L, Hurst S, Balajee SA et al (2017) Detection of mucormycetes and other pathogenic fungi in formalin fixed paraffin embedded and fresh tissues using the extended region of 28S rDNA. Med Mycol 55:385–395. https://doi.org/10.1093/mmy/myw083
Galland P, Lipson ED (1985) Modified action spectra of photogeotropic equilibrium in Phycomyces blakesleeanus mutants with defects in genes madA, madB, madC, and madH. Photochem Photobiol 41:331–335. https://doi.org/10.1111/j.1751-1097.1985.tb03493.x
Gams W (1961) Eine neue Mortierella aus dem zentralalpinen Rothumus. Nova Hedwigia 3:69–71
Gams W (1976) Some new or noteworthy species of Mortierella. Persoonia 9:111–140
Gams W (1977) A key to the species of Mortierella. Persoonia 9:381–391
Gan T, Luo T, Pang K et al (2021) Cryptic terrestrial fungus-like fossils of the early Ediacaran Period. Nat Commun 12:1–12. https://doi.org/10.1038/S41467-021-20975-1
Gardeli C, Athenaki M, Xenopoulos E et al (2017) Lipid production and characterization by Mortierella (Umbelopsis) isabellina cultivated on lignocellulosic sugars. J Appl Microbiol 123:1461–1477. https://doi.org/10.1111/jam.13587
Gaya E, Fernández-Brime S, Vargas R et al (2015) The adaptive radiation of lichen-forming Teloschistaceae is associated with sunscreening pigments and a bark-to-rock substrate shift. PANS 112:11600–11605. https://doi.org/10.1073/pnas.1507072112
Gomes MZ, Lewis RE, Kontoyiannis DP (2011) Mucormycosis caused by unusual mucormycetes, non-Rhizopus, -Mucor, and -Lichtheimia species. Clin Microbiol Rev 24:411–445. https://doi.org/10.1128/CMR.00056-10
Guarro J, Chander J, Alvarez E et al (2011) Apophysomyces variabilis infections in humans. Emerg Infect Dis 17:134–135. https://doi.org/10.3201/eid1701.101139
Gueidan C, Ruibal C, de Hoog G et al (2011) Rock-inhabiting fungi originated during periods of dry climate in the late Devonian and middle Triassic. Fungal Biol 115:987–996. https://doi.org/10.1016/j.funb.2011.04.002
Guo J, Wang H, Liu D et al (2015) Isolation of Cunninghamella bigelovii sp. nov. CGMCC 8094 as a new endophytic oleaginous fungus from Salicornia bigelovii. Mycol Prog 14:1–8. https://doi.org/10.1007/s11557-015-1029-z
Guo LW, Wu YX, Mao ZC et al (2012) Storage rot of dragon fruit caused by Gilbertella persicaria. Plant Dis 96:1826–1826. https://doi.org/10.1094/PDIS-07-12-0635-PDN
Hagem O (1910) Neue untersuchungen über norwegische mucorineen. Annales Mycologici 8:265–286
Hallur V, Prakash H, Sable M et al (2021) Cunninghamella arunalokei a new species of Cunninghamella from India causing disease in an immunocompetent individual. J Fungi 7:670. https://doi.org/10.3390/JOF7080670
Han B, Rombouts FM, Nout MJR (2001) A Chinese fermented soybean food. Int J Food Microbiol 65:1–10. https://doi.org/10.1016/S0168-1605(00)00523-7
He MQ, Zhao RL, Hyde KD et al (2019) Notes, outline and divergence times of Basidiomycota. Fungal Divers 99:105–367. https://doi.org/10.1007/s13225-019-00435-4
Heckman DS, Geiser DM, Eidell BR et al (2001) Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129–1133. https://doi.org/10.1126/science.1061457
Hermet A, Méheust D, Mounier J et al (2012) Molecular systematics in the genus Mucor with special regards to species encountered in cheese. Fungal Biol 116:692–705. https://doi.org/10.1016/j.funbio.2015.06.001
Hesse CN, Torres-Cruz TJ, Tobias TB et al (2016) Ribosomal RNA gene detection and targeted culture of novel nitrogen-responsive fungal taxa from temperate pine forest soil. Mycologia 108:1082–1090
Hesseltine CW (1960) Gilbertella gen. nov. (Mucorales). Bull Torrey Bot Club 87:21–30. https://doi.org/10.2307/2483058
Hesseltine CW, Anderson P (1956) The genus Thamnidium and a study of the formation of its zygospores. Am J Bot 43:696–703. https://doi.org/10.1002/j.1537-2197.1956.tb14434.x
Hesseltine CW, Benjamin CR, Mehrotra BS (1959) The genus Zygorhynchus. Mycologia 51:173–194. https://doi.org/10.1080/00275514.1959.12024811
Hesseltine CW, Ellis JJ (1961) Notes on mucorales, especially Absidia. Mycologia 53:406–426. https://doi.org/10.1080/00275514.1961.12017970
Hesseltine CW, Ellis JJ (1964) The genus Absidia: Gongronella and cylindrical-spored species of Absidia. Mycologia 56:568–601. https://doi.org/10.1080/00275514.1964.12018145
Hesseltine CW, Ellis JJ (1966) Species of Absidia with ovoid sporangiospores 1. Mycologia 58:761–785. https://doi.org/10.1080/00275514.1966.12018369
Hesseltine CW, Fennell DI (1955) The genus Circinella. Mycologia 47:193–212. https://doi.org/10.1080/00275514.1955.12024444
Hibbett DS, Binder M, Bischoff JF et al (2007) A higher-level phylogenetic classification of the fungi. Mycol Res 111:509–547. https://doi.org/10.1016/j.mycres.2007.03.004
Hibbett DS, Taylor JW (2013) Fungal systematics: is a new age of enlightenment at hand? Nat Rev Microbiol 11:129–133. https://doi.org/10.1038/nrmicro2963
Ho HM (1988) The study of Rhizopus stolonifer isolated in Taiwan 2. Morphological study of Rhizopus stolonifer: asexual apparatus and zygosporogenesis. Trans Mycol Soc Rep China 3:73–81
Ho HM (1995a) Notes on two coprophilous species of the genus Circinella (Mucorales) from Taiwan. Fungal Sci 10:23–27
Ho HM (1995b) Zygospore wall in early stage of Rhizopus sexualis (Mucoraceae): smooth or warted? Fungal Sci 10:29–31
Ho HM (2002) Notes on Zygomycetes of Taiwan 2: two Thamnidiaceae (Mucorales) fungi. Fungal Sci 17:87–92
Ho HM, Chang LL (2003) Notes on Zygomycetes of Taiwan 3: two Blakeslea species (Choanephoraceae) new to Taiwan. Taiwania 48:232–238. https://doi.org/10.6165/tai.2003.48(4).232
Ho HM, Chen ZC (1990) Morphological study of Gongronella butleri (Mucorales) from Taiwan. Taiwania 35:259–263. https://doi.org/10.6165/tai.1990.35.259
Ho HM, Chen ZC (1994) Post cleavage transformation of columella in Rhizopus stolonifer (Mucoraceae). Botanical Bulletin of Academia Sinica 35:249–259
Ho HM, Chuang SC, Chen SJ (2004) Notes on Zygomycetes of Taiwan 4: three Absidia species (Mucoraceae). Fungal Sci 19:125–131
Ho HM, Chuang SC, Hsieh CY (2008) Notes on Zygomycetes of Taiwan 6: Chaetocladium brefeldii new to Taiwan. Fungal Sci 23:21–25
Hoffmann K (2010) Identification of the genus Absidia (Mucorales, Zygomycetes): a comprehensive taxonomic revision. In: Gherbawy Y, Voigt K (eds) Molecular identification of fungi, 1st, edn. Springer, Berlin, pp 439–460. https://doi.org/10.1007/978-3-642-05042-8_19
Hoffmann K, Discher S, Voigt K (2007) Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group Mycocladiaceae fam. nov. Mycol Res 111:1169–1183. https://doi.org/10.1016/j.mycres.2007.07.002
Hoffmann K, Pawłowska J, Walther G et al (2013) The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 30:57–76. https://doi.org/10.3767/003158513X666259
Hoffmann K, Telle S, Walther G et al (2009a) Diversity, genotypic identification, ultrastructural and phylogenetic characterization of zygomycetes from different ecological habitats and climatic regions: limitations and utility of nuclear ribosomal DNA barcode markers. In: Gherbawy Y, Mach R, Rai M (eds) Current advances in molecular mycology, 1st, edn. Nova Science Publishers, New York, pp 263–312
Hoffmann K, Voigt K (2009) Absidia parricida plays a dominant role in biotrophic fusion parasitism among mucoralean fungi (Zygomycetes): Lentamyces, a new genus for A. parricida and A. zychae. Plant Biol 11:537–554. https://doi.org/10.1111/j.1438-8677.2008.00145.x
Hoffmann K, Voigt K, Kirk PM (2011) Mortierellomycotina subphyl. nov., based on multi–gene genealogies. Mycotaxon 115:353–363. https://doi.org/10.5248/115.353
Hoffmann K, Walther G, Voigt K (2009b) Mycocladus vs. Lichtheimia: a correction (Lichtheimiaceae fam. nov., Mucorales, Mucoromycotina). Mycol Res 113:277–278
Hong SB, Kim DH, Lee M et al (2012) Zygomycota associated with traditional meju, a fermented soybean starting material for soy sauce and soybean paste. J Microbiol 50:386–393. https://doi.org/10.1007/s12275-012-1437-6
Horn F, Üzüm Z, Möbius N et al (2015) Draft genome sequences of symbiotic and nonsymbiotic Rhizopus microsporus strains CBS 344.29 and ATCC 62417. Genome Announc 3:e01370-e1414. https://doi.org/10.1128/genomeA.01370-14
Hsu TH, Ho HM (2010) Notes on Zygomycetes of Taiwan 8: three new records of Absidia in Taiwan. Fungal Sci 25:5–11
Hsu TH, Ho HM , Chien C (2009) Taxonomic study of Absidia sensu lato in Taiwan. Asian Mycological Congress and 11th International Marine and Freshwater Mycology Symposium Abstract Book, pp 42.
Hu FM, Zheng RY, Chen GQ (1989) A redelimitation of the species of Pilobolus. Mycosystema 2:1–133
Hu J, Zhang Y, Xu Y et al (2019) Gongronella sp. w5 elevates Coprinopsis cinerea laccase production by carbon source syntrophism and secondary metabolite induction. Appl Microbiol Biotechnol 103:411–425. https://doi.org/10.1007/s00253-018-9469-4
Huang J, Zhang J, Zhang X et al (2014) Mucor fragilis as a novel source of the key pharmaceutical agents podophyllotoxin and kaempferol. Pharm Biol 52:237–1243. https://doi.org/10.3109/13880209.2014.885061
Huang R, Gou J, Zhao D et al (2018) Phytotoxicity and anti-phytopathogenic activities of marine-derived fungi and their secondary metabolites. RSC Adv 8:37573–37580. https://doi.org/10.1039/c8ra08047j
Huët MAL, Wong LW, Goh CBS et al (2020) First reported case of Gilbertella persicaria in human stool: outcome of a community study from Segamat, Johor, Malaysia. Braz J Microbiol 51:2067–2075. https://doi.org/10.1007/s42770-020-00323-z
Hurdeal VG, Gentekaki E, Lee HB et al (2021) Mucoralean fungi in Thailand: novel species of Absidia from tropical forest soil. Cryptogamie Mycol 42:39–61. https://doi.org/10.5252/CRYPTOGAMIE-MYCOLOGIE2021V42A4
Hurdeal VG, Jones EBG, de Santiago ALCM et al (2022) Expanding the diversity of mucoralean fungi from northern Thailand: novel Backusella species from soil. Phytotaxa 559:275–284. https://doi.org/10.11646/phytotaxa.559.3.5
Hyde KD, Hongsanan S, Jeewon R et al (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 80:1–270. https://doi.org/10.1007/s13225-016-0373-x
Hyde KD, Maharachchikumbura SS, Hongsanan S et al (2017) The ranking of fungi: a tribute to David L. Hawksworth on his 70th birthday. Fungal Divers 84:1–23. https://doi.org/10.1007/s13225-017-0383-3
Hyde KD, Suwannarach N, Jayawardena RS et al (2021) Mycosphere notes 325–344 – novel species and records of fungal taxa from around the world. Mycosphores 12:1101–1156. https://doi.org/10.5943/mycosphere/12/1/14
Irshad M, Nasir N, Hashmi UH et al (2020) Invasive pulmonary infection by Syncephalastrum species: two case reports and review of literature. IDCases 21:e00913. https://doi.org/10.1016/j.idcr.2020.e00913
James TY, Kauff F, Schoch CL et al (2006) Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818–822. https://doi.org/10.1038/nature05110
James TY, Pelin A, Bonen L et al (2013) Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol 23:1548–1553. https://doi.org/10.1016/j.cub.2013.06.057
Jia H, Zhang M, Weng Y et al (2021) Degradation of polylactic acid/polybutylene adipate-co-terephthalate by coculture of Pseudomonas mendocina and Actinomucor elegans. J Hazard Mater 403:123679. https://doi.org/10.1016/j.jhazmat.2020.123679
Ju X, Zhang MZ, Zhao H et al (2020) Genomic SNPs reveal population structure of Rhizopus arrhizus. Mycosystema 39:2285–2303. https://doi.org/10.13346/j.mycosystema.200105
Kaitera J, Henttonen HM, Müller MM (2019) Fungal species associated with butt rot of mature Scots pine and Norway spruce in northern boreal forests of Northern Ostrobothnia and Kainuu in Finland. Eur J Plant Pathol 154:541–554. https://doi.org/10.1007/s10658-019-01678-2
Kamei K (2000) Animal models of Zygomycosis—Absidia, Rhizopus, Rhizomucor, and Cunninghamella. Mycopathologia 152:5–13. https://doi.org/10.1023/A:1011900630987
Kanouse BB (1936) Studies of two species of Endogone in culture. Mycologia 28:47–62. https://doi.org/10.1080/00275514.1936.12017117
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. https://doi.org/10.1093/bib/bbx108
Kearse M, Moir R, Wilson A et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Khale A, Deshpande MV (1992) Dimorphism in Benjaminiella poitrasii: cell wall chemistry of parent and two stable yeast mutants. Antonie Van Leeuwenhoek 62:299–307. https://doi.org/10.1007/BF00572598
Khan MA, Yang J, Hussain SA et al (2019) Genetic modification of Mucor circinelloides to construct stearidonic acid producing cell factory. Int J Mol Sci 20:1683. https://doi.org/10.3390/ijms20071683
Khuna S, Suwannarach N, Kumla J et al (2019) Apophysomyces thailandensis (Mucorales, Mucoromycota), a new species isolated from soil in northern Thailand and its solubilization of non-soluble minerals. MycoKeys 45:75–92. https://doi.org/10.3897/mycokeys.45.30813
Kirk PM (1984) A monograph of the Choanephoraceae. Mycol Pap 152:1–61
Kirk PM (1989) A new species of Benjaminiella (Mucorales: Mycotyphaceae). Mycotaxonomy 35:121–125
Kirk PM, Benny GL (1980) Genus Utharomyces Boedijn (Pilobolaceae: Zygomycetes). Trans Br Mycol Soc 75:123–131. https://doi.org/10.1016/S0007-1536(80)80202-6
Kitahata S, Ishikawa H, Miyata T et al (1989) Production of rubusoside derivatives by transgalactosylation of various α-galactosidases. Agric Biol Chem 53:2929–2934. https://doi.org/10.1271/bbb1961.53.2929
Kitz DJ, Embree RW Jr, Cazin J (1980) Radiomyces a genus in the mucorales pathogenic for mice. Sabouraudia 18:115–121. https://doi.org/10.1080/00362178085380191
Kosa G, Zimmermann B, Kohler A et al (2018) High-throughput screening of Mucoromycota fungi for production of low- and high-value lipids. Biotechnol Biofuels 11:1–17. https://doi.org/10.1186/s13068-018-1070-7
Kovacs R, Sundberg W (1999) Syzygites megalocarpus (Mucorales, Zygomycetes) in Illinois. Trans Illinois State Acad Sci 92:181–190
Krings M, Harper CJ, Taylor EL (2018) Fungi and fungal interactions in the Rhynie chert: a review of the evidence, with the description of Perexiflasca tayloriana gen. et sp. nov. Philos Trans R Soc B 373:500. https://doi.org/10.1098/rstb.2016.0500
Krings M, Taylor TN, Dotzler N (2011) The fossil record of the Peronosporomycetes (Oomycota). Mycologia 103:445–457. https://doi.org/10.3852/10-278
Kristanti RA, Zubir MMFA, Hadibarata T (2016) Biotransformation studies of cresol red by Absidia spinosa M15. J Environ Manage 172:107–111. https://doi.org/10.1016/j.jenvman.2015.11.017
Kumar P, Satyanarayana T (2007) Economical glucoamylase production by alginate-immobilized Thermomucor indicae-seudaticae in cane molasses medium. Lett Appl Microbiol 45:392–397. https://doi.org/10.1111/j.1472-765X.2007.02201.x
Kwon-Chung KJ (2012) Taxonomy of fungi causing mucormycosis and entomophthoramycosis (zygomycosis) and nomenclature of the disease: molecular mycologic perspectives. Clin Infect Dis 54(Suppl 1):S8–S15. https://doi.org/10.1093/cid/cir864
Labuda R, Bernreiter A, Hochenauer D et al (2019) Saksenaea dorisiae sp. nov., a new opportunistic pathogenic fungus from Europe. Int J Microbiol. https://doi.org/10.1155/2019/6253829
Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. https://doi.org/10.1093/bioinformatics/btu531
Layios N, Canivet J, Baron F et al (2014) Mortierella wolfii–associated invasive disease. Emerg Infect Dis 20:1591–1592. https://doi.org/10.3201/eid2009.140469
Lee SC, Billmyre RB, Li A et al (2014) Analysis of a food-borne fungal pathogen outbreak: virulence and genome of a Mucor circinelloides isolate from yogurt. MBio. https://doi.org/10.1128/mBio.01390-14
Lee SH, Nguyen TTT, Lee HB (2018) Isolation and characterization of two rare Mucoralean species with specific habitats. Mycobiology 46:205–214. https://doi.org/10.1080/12298093.2018.1509513
Li F, Zhang S, Wang Y et al (2020) Rare fungus, Mortierella capitata, promotes crop growth by stimulating primary metabolisms related genes and reshaping rhizosphere bacterial community. Soil Biol Biochem 151:108017. https://doi.org/10.1016/j.soilbio.2020.108017
Li G, Fang X, Su F et al (2018) Enhancing the thermostability of Rhizomucor miehei lipase with a limited screening library by rational-design point mutations and disulfide bonds. Appl Environ Microbiol 84:e02129-e2217. https://doi.org/10.1128/AEM.02129-17
Li GJ, Hyde KD, Zhao RL et al (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 78:1–237. https://doi.org/10.1007/s13225-016-0366-9
Li S, Han R, Zhang H et al (2020) Apophysomyces jiangsuensis sp. nov., a salt tolerant and phosphate-solubilizing fungus from the Tidelands of Jiangsu Province of China. Microorganisms 8:1868. https://doi.org/10.3390/MICROORGANISMS8121868
Linde J, Schwartze V, Binder U et al (2014) De novo whole-genome sequence and genome annotation of Lichtheimia ramosa. Genome Announcements. https://doi.org/10.1128/genomeA.00888-14
Linnemann G (1941) Die Mucorineen-Gattung Mortierella Coemans. Pflanzenforschung 23:1–64
Liu F, Ma ZY, Hou LW et al (2022a) Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud Mycol 101:1–56. https://doi.org/10.3114/sim.2022.101.01
Liu XY (2004) Pirella (Thamnidiaceae, Mucorales), a new record genus for China. Mycosystema 23:301–302. https://doi.org/10.3969/j.issn.1672-6472.2004.02.023
Liu XY, Huang H, Zheng RY (2001) Relationships within Cunninghamella based on sequence analysis of ITS rDNA. Mycotaxon 80:77–95
Liu XY, Huang H, Zheng RY (2008) Delimitation of Rhizopus varieties based on IGS rDNA sequences. Sydowia 60:93–112
Liu XY, Voigt K (2010) Molecular characters of zygomycetous fungi. In: Gherbawy Y, Voigt K (eds) Molecular identification of fungi. Springer, Berlin, pp 461–488. https://doi.org/10.1007/978-3-642-05042-8_20
Liu XY, Wang Y, Zheng RY (2012) Molecular phylogeny of Pilaira (Mucorales, Zygomycetes) inferred from ITS rDNA and pyrG sequences. Sydowia 64:55–66
Liu XY, Zheng RY (2015) New taxa of Ambomucor (Mucorales, Mucoromycotina) from China. Mycotaxon 130:165–171. https://doi.org/10.5248/130.165
Liu ZB, Wu YD, Zhao H et al (2022) Outline, divergence times, and phylogeneticanalyses of Trechisporales (Agaricomycetes, Basidiomycota). Front Microbiol 13:818358. https://doi.org/10.3389/fmicb.2022.818358
Loh L, Nawawi A, Kuthubutheen A (2001) Mucoraceous fungi from Malaysia. Dissertation, University of Malaya
Lu JM, Yu RC, Chou CC (1996) Purification and some properties of glutaminase from Actinomucor taiwanensis, starter of sufu. J Sci Food Agr 70:509–514. https://doi.org/10.1002/(SICI)1097-0010(199604)70:4%3c509::AID-JSFA532%3e3.0.CO;2-7
Lucet A, Costantin J (1900) Rhizomucor parasiticus. EspeA ce pathogeAne delhomme. Rev GeAneArale Bot 12:89–98
Lucking R, Huhndorf S, Pfister DH et al (2009) Fungi evolved right on track. Mycologia 101:810–822. https://doi.org/10.3852/09-016
Lukacs B, Papp T, Nyilasi I et al (2004) Differentiation of Rhizomucor species on the basis of their different sensitivities to lovastatin. J Clin Microbiol 42:5400–5402. https://doi.org/10.1128/JCM.42.11.5400-5402.2004
Luo W, Xue C, Zhao Y et al (2020) Blakeslea trispora photoreceptors: identification and functional analysis. Appl Environ Microbiol 86:e02962-e3019. https://doi.org/10.1128/AEM.02962-19
Mahalaxmi I, Jayaramayya K, Venkatesan D et al (2021) Mucormycosis: an opportunistic pathogen during COVID–19. Environ Res. https://doi.org/10.1016/j.envres.2021.111643
Mahmud A, Lee R, Munfus-McCray D et al (2012) Actinomucor elegans as an emerging cause of mucormycosis. J Clin Microbiol 50:1092–1095. https://doi.org/10.1128/JCM.05338-11
Malar CM, Krüger M, Krüger C et al (2021) The genome of Geosiphon pyriformis reveals ancestral traits linked to the emergence of the arbuscular mycorrhizal symbiosis. Curr Biol 31:1570–1577. https://doi.org/10.1016/j.cub.2021.01.058
Malar CM, Wang Y, Stajich JE et al (2022) Early branching arbuscular mycorrhizal fungus Paraglomus occultum carries a small and repeat-poor genome compared to relatives in the Glomeromycotina. Microb Genomics 8:810. https://doi.org/10.1099/mgen.0.000810
Martínez-Herrera E, Frías-De-León MG, Julián-Castrejón A et al (2020) Rhino-orbital mucormycosis due to Apophysomyces ossiformis in a patient with diabetes mellitus: a case report. BMC Infect Dis 20:1–4. https://doi.org/10.1186/s12879-020-05337-4
Martins EDS, Gomes E, da Silva R et al (2019) Production of cellulases by Thermomucor indicae-seudaticae: characterization of a thermophilic β-glucosidase. Prep Biochem Biotech 49:830–836. https://doi.org/10.1080/10826068.2019.1625060
Martins MR, Santos C, Soares C et al (2020) Gongronella eborensis sp. nov., from vineyard soil of Alentejo (Portugal). Int J Syst Evol Microsc 70:3475–3482. https://doi.org/10.1099/ijsem.0.004201
McGee PA (1996) The Australian zygomycetous mycorrhizal fungi: the genus Densospora gen. nov. Aust Syst Bot 9:329–336. https://doi.org/10.1071/SB9960329
Mehrotra BS, Baijal U, Mehrotra BR (1963) Two new species of Moriterella from India. Mycologia 55:289–296. https://doi.org/10.11646/zootaxa.4742.1.3
Mehrotra BS, Mehrotra BR (1969) Two new species of Mucor from India 4. Sydowia 23:183–185
Merheb-Dini C, Garcia GAC, Penna ALB et al (2012) Use of a new milk-clotting protease from Thermomucor indicae-seudaticae N31 as coagulant and changes during ripening of Prato cheese. Food Chem 130:859–865. https://doi.org/10.1016/j.foodchem.2011.07.105
Meyer W, Gams W (2003) Delimitation of Umbelopsis (Mucorales, Umbelopsidaceae fam. nov.) based on ITS sequence and RFLP data. Mycol Res 107:339–350. https://doi.org/10.1017/s0953756203007226
Mirza J, Khan S, Begum S et al (1979) Mucorales of Pakistan. University of Agriculture, Faisalabad, pp 1–183
Misra PC (1975) A new species of Syncephalastrum. Mycotaxon 3:51–54
Misra PC, Srivastava KJ, Lata K (1979) Apophysomyces, a new genus of the Mucorales. Mycotaxon 8:377–382. https://doi.org/10.1080/00275514.1953.12024280
Mondo SJ, Dannebaum RO, Kuo RC et al (2017) Widespread adenine N6-methylation of active genes in fungi. Nat Genet 49:964–968. https://doi.org/10.1038/ng.3859
Moreau F (1952) Les champignons, physiologie, morphologie développement ét systematique. Botany, Paris
Morin E, Miyauchi S, San Clemente H et al (2019) Comparative genomics of Rhizophagus irregularis, R. cerebriforme, R. diaphanus and Gigaspora rosea highlights specific genetic features in Glomeromycotina. New Phytol 222:1584–1598. https://doi.org/10.1111/nph.15687
Morin-Sardin S, Nodet P, Coton E et al (2017) Mucor: a Janus-faced fungal genus with human health impact and industrial applications. Fungal Biol Rev 31:12–32. https://doi.org/10.1016/j.fbr.2016.11.002
Munday JS, Laven RA, Orbell GMB et al (2006) Meningoencephalitis in an adult cow due to Mortierella wolfii. J Vet Diagn Invest 18:619–622. https://doi.org/10.1177/104063870601800620
Munday JS, Wolfe AG, Lawrence KE et al (2010) Disseminated Mortierella wolfii infection in a neonatal calf. New Zeal Vet J 58:62–63. https://doi.org/10.1080/00480169.2010.65062
Nguyen MH, Kaul D, Muto C et al (2020) Genetic diversity of clinical and environmental Mucorales isolates obtained from an investigation of mucormycosis cases among solid organ transplant recipients. Microbial Genomics 6:mgen000473. https://doi.org/10.1099/mgen.0.000473
Nguyen TTT, Voigt K, de Santiago ALCMA et al (2021) Discovery of novel Backusella (Backusellaceae, Mucorales) isolated from invertebrates and toads in Cheongyang. Korea J Fungi 7:513. https://doi.org/10.3390/jof7070513
Nguyen TTT, Lee HB (2022) Discovery of three new Mucor species associated with cricket insects in Korea. J Fungi 8:601. https://doi.org/10.3390/jof8060601
Nie Y, Cai Y, Gao Y et al (2020a) Three new species of Conidiobolus sensu stricto from plant debris in eastern China. MycoKeys 73:133–149. https://doi.org/10.3897/mycokeys.73.56905
Nie Y, Yu DS, Wang CF et al (2020b) A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): four clades including three new genera. MycoKeys 66:55–81. https://doi.org/10.3897/mycokeys.66.46575
Nilsson RH, Hyde KD, Pawłowska J et al (2014) Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Divers 67:11–19. https://doi.org/10.1007/s13225-014-0291-8
O’Donnell K, Lutzoni FM, Ward TJ et al (2001) Evolutionary relationships among mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia 93:286–297. https://doi.org/10.1111/nph.15687
Oehl F, da Sliva GA, Goto BT et al (2011a) Glomeromycota: two new classes and a new order. Mycotaxon 116:365–379. https://doi.org/10.5248/116.365
Oehl F, da Sliva GA, Goto BT et al (2011) Glomeromycota: three new genera and glomoid species reorganized. Mycotaxon 116:75–120
Oehl F, Sieverding E, Palenzuela J et al (2011c) Advances in Glomeromycota taxonomy and classification. IMA Fungus 2:191–199. https://doi.org/10.5598/imafungus.2011.02.02.10
Overton BE (1997) Notes on the genus Spinellus with comparisons to other Zygomycetes found in fungicolous associations with Basidiomycetes. Dissertation, Southern Illinois University at Carbondale
Ozimek E, Hanaka A (2021) Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agriculture 11:7. https://doi.org/10.3390/AGRICULTURE11010007
Page RM (1962) Light and the asexual reproduction of Pilobolus. Science 138:1238–1245. https://doi.org/10.1126/science.138.3546.1238
Page RM (1964) Sporangium discharge in Pilobolus: a photographic study. Science 146:925–927. https://doi.org/10.1126/science.146.3646.925
Page RM, Kennedy D (1964) Studies on the velocity of discharged sporangia of Pilobolus kleinii. Mycologia 56:363–368. https://doi.org/10.1080/00275514.1964.12018119
Panthee S, Hamamoto H, Nishiyama Y et al (2021) Novel pathogenic Mucorales identified using the silkworm infection model. J Fungi 7:995. https://doi.org/10.3390/JOF7110995
Papp T, Nagy G, Csernetics A et al (2009) Beta-carotene production by mucoralean fungi. J Eng Ann Fac Eng Hunedoara 7:173–176
Pavlovic MD, Bulajic N (2006) Great toenail onychomycosis caused by Syncephalastrum racemosum. Dermatol Onl J 12:7. https://doi.org/10.5070/D3794644t6
Pei KQ (2000) A new variety of Mucor variosporus and the validation of M. luteus Linnemann and M. variosporus Schipper. Mycosystema 19:10–12. https://doi.org/10.13346/j.mycosystema.2000.01.004
Pei KQ (2000b) Three new records of Mucor from China. Mycosystema 19:563–565. https://doi.org/10.3969/j.issn.1672-6472.2000.04.024
Pereira WES, da Silva RR, de Amo GS et al (2020) A collagenolytic aspartic protease from Thermomucor indicae-seudaticae expressed in Escherichia coli and Pichia pastoris. Appl Biochem Biotechn 191:1258–1270. https://doi.org/10.1007/s12010-020-03292-z
Pertea G, Pertea M (2020) GFF utilities: GffRead and GffCompare. F1000Research 9:304. https://doi.org/10.12688/f1000research.23297.1
Petkovits T, Nagy LG, Hoffmann K et al (2011) Data partitions, Bayesian analysis and phylogeny of the zygomycetous fungal family Mortierellaceae, inferred from nuclear ribosomal DNA sequences. PLoS ONE 6:e27507. https://doi.org/10.1371/journal.pone.0027507
Phookamsak R, Hyde KD, Jeewon R et al (2019) Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers 95:1–273. https://doi.org/10.1007/s13225-019-00421-w
Pinho DB, Pereira OL, Soares DJ (2014) First report of Gilbertella persicaria as the cause of soft rot of fruit of Syzygium cumini. Australas Plant Dis 9:143. https://doi.org/10.1007/s13314-014-0143-0
Prakash H, Rudramurthy SM, Gandham PS et al (2017) Apophysomyces variabilis: draft genome sequence and comparison of predictive virulence determinants with other medically important Mucorales. BMC Genomics 18:1–13. https://doi.org/10.1186/s12864-017-4136-1
Qin D, Wang L, Han M et al (2018) Effects of an endophytic fungus Umbelopsis dimorpha on the secondary metabolites of host–plant Kadsura angustifolia. Front Microbiol 9:2845. https://doi.org/10.3389/fmicb.2018.02845
Rafhaella T, Cordeiro L, Thuong T et al (2020) Two new species of the industrially relevant genus Absidia (Mucorales) from soil of the Brazilian Atlantic Forest. Acta Bot Bras 34:1–10. https://doi.org/10.1590/0102-33062020abb0040
Rall G, Solheim W (1964) A variety of Absidia isolated from Comandra pallida. Mycologia 56:99–102. https://doi.org/10.1080/00275514.1964.12018086
Rambaut A, Drummond A (2013) Tracer v1. 5. http://beast.bio.ed.ac.uk/Tracer
Rao CY, Kurukularatne C, Garcia-Diaz JB et al (2007) Implications of detecting the mold Syncephalastrum in clinical specimens of new orleans residents after Hurricanes Katrina and Rita. J Occup Environ Med 49:411–416. https://doi.org/10.1097/JOM.0b013e31803b94f9
Rashmi M, Kushveer JS, Sarma VV (2019) A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10:798–1079. https://doi.org/10.5943/mycosphere/10/1/19
Redecker D, Kodner R, Graham LE (2000) Glomalean fungi from the Ordovician. Science 289:1920–1921. https://doi.org/10.1126/science.289.5486.1920
Ribaldi M (1952) Sopra un interessante Zigomicete terricola: Gongronella urceolifera n. gen., n. sp. Riv Biol Gen, NS 44:157–166
Ridgway R (1912) Color standards and color nomenclature. D. C., The author, Washington
Robinson BE, Stark MT, Pope TL et al (1990) Cunninghamella bertholletiae: an unusual agent of zygomycosis. South Med J 83:1088–1091
Romero-Olivares AL, Meléndrez-Carballo G, Lago-Lestón A et al (2019) Soil metatranscriptomes under long-term experimental warming and drying: fungi allocate resources to cell metabolic maintenance rather than decay. Front Microbiol 10:1914. https://doi.org/10.3389/fmicb.2019.01914
Ronquist F, Teslenko M, Van Der Mark P et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Saksena S (1953) A new genus of the Mucorales. Mycologia 45:426–436. https://doi.org/10.1080/00275514.1953.12024280
Sanchez R, Serra F, Tarraga J et al (2011) Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res 39:W470–W474. https://doi.org/10.1093/nar/gkr408
Sandmann G (2021) Diversity and origin of carotenoid biosynthesis: its history of coevolution towards plant photosynthesis. New Phytol 232:479–493. https://doi.org/10.1111/NPH.17655
Saroj A, Kumar A, Qamar N et al (2012) First report of wet rot of Withania somnifera caused by Choanephora cucurbitarum in India. Plant Dis 96:293–293. https://doi.org/10.1094/PDIS-09-11-0801
Schipper MAA (1973) A study on variability in Mucor hiemalis and related species. Stud Mycol 4:1–40
Schipper MAA (1976) On Mucor circinelloides, Mucor racemosus and related species. Stud Mycol 12:1–40. https://doi.org/10.1016/S0007-1536(76)80201-X
Schipper MAA (1978a) On certain species of Mucor with a key to all accepted species. Stud Mycol 17:1–52
Schipper MAA (1978b) On the genera Rhizomucor and Parasitella. Stud Mycol 17:53–71
Schipper MAA (1979) Thermomucor (Mucorales). Antonie Van Leeuwenhoek 45:275–280. https://doi.org/10.1007/BF00418590
Schipper MAA (1986a) Hyphomucor, an new genus in the Mucorales for Mucor assamensis. Mycotaxon 27:83–86
Schipper MAA (1986b) Notes on Zygorhynchus species. Persoonia 13:97–105
Schipper MAA, Samson RA (1994) Miscellaneous notes on Mucoraceae. Mycotaxon 50:475–491
Schipper MAA (1990) Notes on Mucorales—1. Observations on Absidia. Persoonia 14:133–149
Schoch CL, Seifert KA, Huhndorf S et al (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. PANS 109:6241–6246. https://doi.org/10.1073/pnas.1117018109
Schüßler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421. https://doi.org/10.1017/S0953756201005196
Schulz BE, Kraepelin G, Hinkelmann W (1974) Factors affecting dimorphism in Mycotypha (Mucorales): a correlation with the fermentation/respiration equilibrium. Microbiology 82:1–13. https://doi.org/10.1099/00221287-82-1-1
Schwartze VU, Jacobsen ID (2014) Mucormycoses caused by Lichtheimia species. Mycoses 57:73–78. https://doi.org/10.1111/myc.12239
Schwartze VU, Winter S, Shelest E et al (2014) Gene expansion shapes genome architecture in the human pathogen Lichtheimia corymbifera: an evolutionary genomics analysis in the ancient terrestrial mucorales (Mucoromycotina). PLoS Genetic 10:e1004496. https://doi.org/10.1371/journal.pgen.1004496
Sepuri A, Vidyavathi M (2008) Cunninghamella—a microbial model for drug metabolism studies—a review. Biotechnol Adv 27:16–29. https://doi.org/10.1111/myc.12239
Shah AM, Mohamed H, Zhang Z et al (2021) Isolation, characterization and fatty acid analysis of Gilbertella persicaria DSR1: a potential new source of high value single-cell oil. Biomass Bioenerg 151:106156. https://doi.org/10.1016/J.BIOMBIOE.2021.106156
Shah NN, Khan Z, Ahad H et al (2022) Mucormycosis an added burden to Covid-19 Patients: an in-depth systematic review. J Infect Public Heath 15:1299–1314. https://doi.org/10.1016/j.jiph.2022.10.011
Shanor L, Poitras AW, Benjamin RK (1950) A new genus of the Choanephoraceae. Mycologia 42:271–278. https://doi.org/10.1080/00275514.1950.12017831
Shipton W, Schipper MAA (1975) Halteromyces, a new genus in the Mucorales. Antonie Van Leeuwenhoek 41:337–342. https://doi.org/10.1007/BF02565068
Smith ME, Gryganskyi A, Bonito G et al (2013) Phylogenetic analysis of the genus Modicella reveals an independent evolutionary origin of sporocarp-forming fungi in the Mortierellales. Fungal Genet Biol 61:61–68. https://doi.org/10.1016/j.fgb.2013.10.001
Spatafora JW, Chang Y, Benny GL et al (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108:1028–1046. https://doi.org/10.3852/16-042
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stanke M, Waack S (2003) Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19:215–225. https://doi.org/10.1093/bioinformatics/btg1080
Subrahmanyam A, Lakshmi BV (1993) Thermomucor indicae-seudaticae on human skin. Mycoses 36:201–202. https://doi.org/10.1111/j.1439-0507.1993.tb00750.x
Subrahmanyam A, Mehrotra B, Thirumalachar M (1977) Thermomucor, a new genus of Mucorales. Georgia J Sci 35:1–4
Sugiyama M, Tokumasu S, Gams W (2003) Umbelopsis gibberispora sp. nov. from Japanese leaf litter and a clarification of Micromucor ramannianus var. angulisporus. Mycoscience 44:217–226. https://doi.org/10.1007/s10267-003-0105-4
Takeda I, Tamano K, Yamane N et al (2014) Genome sequence of the Mucoromycotina fungus Umbelopsis isabellina, an effective producer of lipids. Genome Announc 2:e00071-e114. https://doi.org/10.1128/genomeA.00071-14
Tan SC, Tan TK, Wong SM et al (1996) The chitosan yield of zygomycetes at their optimum harvesting time. Carbohyd Polym 30:239–242. https://doi.org/10.1016/S0144-8617(96)00052-5
Taylor AF, Alexander I (2005) The ectomycorrhizal symbiosis: life in the real world. Mycologist 19:102–112. https://doi.org/10.1017/S0269-915X(05)00303-4
Taylor JW, Jacobson DJ, Kroken S et al (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32. https://doi.org/10.1006/fgbi.2000.1228
Taylor TN, Krings M, Taylor EL (2014) Fossil fungi. Academic Press, London, pp 1–383
Tedersoo L, Sánchez-Ramírez S, Kõljalg U et al (2018) High-level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Divers 90:135–159. https://doi.org/10.1007/s13225-018-0401-0
Telagathoti A, Probst M, Peintner U (2021) Habitat, snow-cover and soil pH, affect the distribution and diversity of Mortierellaceae species and their associations to Bacteria. Front Microbiol 12:1817. https://doi.org/10.3389/FMICB.2021.669784
Thaxter R (1914) New or peculiar Zygomycetes. 3: Blakeslea, Dissophora, and Haplosporangium, nova genera. Bot Gaz 58:353–366
Torres-Cruz TJ, Billingsley Tobias TL, Almatruk M et al (2017) Bifiguratus adelaidae, gen. et sp. nov., a new member of Mucoromycotina in endophytic and soil-dwelling habitats. Mycologia 109:363–378. https://doi.org/10.1080/00275514.2017.1364958
Trachuk P, Szymczak WA, Muscarella P et al (2018) A case of invasive gastrointestinal Mycotypha infection in a patient with neutropenia. Case Rep Infect Dis 2018:5864175. https://doi.org/10.1155/2018/5864175
Trobisch A, Marterer R, Gorkiewicz G et al (2020) Invasive mucormycosis during treatment for acute lymphoblastic leukaemia-successful management of two life-threatening diseases. Support Care Cancer 28:2157–2161. https://doi.org/10.1007/s00520-019-04962-3
Tsai T, Hammond L, Rinaldi M et al (1997) Cokeromyces recurvatus infection in a bone marrow transplant recipient. Bone Marrow Transpl 19:301–302
Tully CC, Romanelli AM, Sutton DA et al (2009) Fatal Actinomucor elegans var. kuwaitiensis infection following combat trauma. J Clin Microbiol 47:3394–3399. https://doi.org/10.1128/JCM.00797-09
Turland NJ, Wiersema JH, Barrie FR et al (2018) International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. Glashütten: Koeltz Botanical Books. https://doi.org/10.12705/Code.2018
Upadhyay H (1970) Soil fungi from north-east and north Brazil 8. Persoonia 6:111–117
Upadhyay H (1973) Helicostylum and Thamnostylum (Mucorales). Mycologia 65:733–751. https://doi.org/10.1080/00275514.1973.12019489
Urquhart AS, Coulon PM, Idnurm A (2017) Pilaira australis sp. nov. Mucorales, Mucoromycota isolated from emu faeces in Australia. Phytotaxa 329:277–283. https://doi.org/10.11646/phytotaxa.329.3.9
Urquhart AS, Douch J, Heafield T et al (2021) Diversity of Backusella (Mucoromycotina) in south-eastern Australia revealed through polyphasic taxonomy. Persoonia 46:1–25. https://doi.org/10.3767/persoonia.2021.46.01
Urquhart AS, Idnurm A (2020) Syncephalastrum contaminatum, a new species in the Mucorales from Australia. Mycoscience 61:111–115. https://doi.org/10.1016/j.myc.2020.02.003
van Tieghem P (1876) Troisieme memoire sur les Mucorinees. Ann Sci Nat Bot Ser 6(4):312–398
Vandepol N, Liber J, Desiro A et al (2020) Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers 104:267–289. https://doi.org/10.1007/s13225-020-00455-5
Viriato A (2008) Pilobolus species found on herbivore dung from the São Paulo Zoological Park, Brazil. Acta Bot Bras 22:614–620. https://doi.org/10.1590/S0102-33062008000300002
Voglmayr H, Krisai-Greilhuber I (1996) Dicranophora fulva, a rare mucoraceous fungus growing on boletes. Mycol Res 100:583–590. https://doi.org/10.1016/S0953-7562(96)80012-8
Voigt K, James TY, Kirk PM et al (2021) Early-diverging fungal phyla: taxonomy, species concept, ecology, distribution, anthropogenic impact, and novel phylogenetic proposals. Fungal Divers 109:59–98. https://doi.org/10.1007/s13225-021-00480-y
Voigt K, Kirk PM (2015) Index Fungorum no. 245. ISSN 2049–2375. Index Fungorum http://www.indexfungorum.org/Publications/Index%20Fungorum%20no.245.pdf.
Voigt K, Wöstemeyer J (2001) Phylogeny and origin of 82 zygomycetes from all 54 genera of the Mucorales and Mortierellales based on combined analysis of actin and translation elongation factor EF-1α genes. Gene 270:113–120. https://doi.org/10.1016/S0378-1119(01)00464-4
von Arx JA (1970) The genera of fungi sporulating in pure culture. pp 1–288. https://doi.org/10.1002/fedr.19820930124
von Arx JA (1982) On Mucoraceae s. str. and other families of the Mucorales. Sydowia 35:10–26
Wagner L, Stielow B, Hoffmann K et al (2013) A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA. Persoonia 30:77–93. https://doi.org/10.3767/003158513X666268
Wagner L, Stielow JB, de Hoog GS et al (2020) A new species concept for the clinically relevant Mucor circinelloides complex. Persoonia 44:67–97. https://doi.org/10.3767/persoonia.2020.44.03
Walsh E, Luo J, Khiste S et al (2021) Pygmaeomycetaceae, a new root-associated family in Mucoromycotina from the pygmy pine plains. Mycologia 113:134–145. https://doi.org/10.1080/00275514.2020.1803649
Walther G, Pawłowska J, Alastruey-Izquierdo et al (2013) DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia 30:11–47. https://doi.org/10.3767/003158513X665070
Wanasinghe DN, Phukhamsakda C, Hyde KD et al (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers 89:1–236. https://doi.org/10.1007/s13225-018-0395-7
Wang K, Chen SL, Dai YC et al (2021) Overview of China’s nomenclature novelties of fungi in the new century (2000–2020). Mycosystema 40:822–833. https://doi.org/10.13346/j.mycosystema.210064
Wang XW, Jiang JH, Liu SL et al (2021b) Species diversification of the coniferous pathogenic fungal genus Coniferiporia (Hymenochaetales, Basidiomycota) in association with its biogeography and host plants. Phytopathology. https://doi.org/10.1094/PHYTO-05-21-0181-R
Wang Y, White MM, Kvist S et al (2016) Genome-wide survey of gut fungi (Harpellales) reveals the first horizontally transferred ubiquitin gene from a mosquito host. Mol Biol Evol 33:2544–2554. https://doi.org/10.1093/molbev/msw126
Wang YJ, Zhao T, Wu WY et al (2022) Cunninghamella verrucosa sp. nov. (Mucorales, Mucoromycota) from Guangdong Province in China. Phytotaxa 560:274–284. https://doi.org/10.11646/phytotaxa.560.3.2
Wang YN, Liu XY, Zheng RY (2013) Four new species records of Umbelopsis (Mucoromycotina) from China. J Mycol 2013:970216. https://doi.org/10.1155/2013/970216
Wang YN, Liu XY, Zheng RY (2014) Umbelopsis changbaiensis sp. nov. from China and the typification of Mortierella vinacea. Mycol Prog 13:657–669. https://doi.org/10.1007/s11557-013-0948-9
Wang YN, Liu XY, Zheng RY (2015) Umbelopsis longicollis comb. nov. and the synonymy of U. roseonana and U. versiformis with U. nana. Mycologia 107:1023–1032. https://doi.org/10.3852/14-339
Wang YN, Liu XY, Zheng RY (2022b) The Umbelopsis ramanniana sensu lato consists of five cryptic species. J Fungi 8:895. https://doi.org/10.3390/jof8090895
Warcup JH (1990) Taxonomy, culture and mycorrhizal associations of some zygosporic Endogonaceae. Mycol Res 94:173–178
Wei F, Hong Y, Liu J et al (2010) Gongronella sp induces overproduction of laccase in Panus rudis. J Basic Microb 50:98–103. https://doi.org/10.1002/jobm.200900155
White TJ, Bruns T, Lee S et al (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications, (ed) Innis, MA, Gelfand DH, Sninsky JJ, and White TJ. Academ- ic Press Inc, New York, pp 315–322
Wijayawardene NN, Hyde KD, Al-Ani LKT et al (2020) Outline of fungi and fungus-like taxa. Mycosphere 11:1060–1456. https://doi.org/10.5943/mycosphere/11/1/8
Wijayawardene NN, Pawłowska J, Letcher PM et al (2018) Notes for genera: basal clades of fungi (including Aphelidiomycota, Basidiobolomycota, Blastocladiomycota, Calcarisporiellomycota, Caulochytriomycota, Chytridiomycota, Entomophthoromycota, Glomeromycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota, Mucoromycota, Neocallimastigomycota, Olpidiomycota, Rozellomycota and Zoopagomycota). Fungal Divers 92:43–129. https://doi.org/10.1007/s13225-018-0409-5
Woodbury N, Gries G (2013) Firebrats, Thermobia domestica, aggregate in response to the microbes Enterobacter cloacae and Mycotypha microspora. Entomol Exp Appl 147:154–159. https://doi.org/10.1111/eea.12054
Wu F, Yuan HS, Zhou LW et al (2020) Polypore diversity in South China. Mycosystema 39:653–682. https://doi.org/10.13346/j.mycosystema.200087
Wu F, Zhou LW, Vlasák J et al (2022) Global diversity and systematics of Hymenochaetaceae with poroid hymenophore. Fungal Divers 113:1–192. https://doi.org/10.1007/s13225-021-00496-4
Wu CG, Lin SJ (1997) Endogonales in Taiwan: a new genus with unizygosporic sporocarps and a hyphal mantle. Mycotaxon 64:179–188
Xess I, Mohapatra S, Shivaprakash MR et al (2012) Evidence implicating Thamnostylum lucknowense as an etiological agent of rhino-orbital mucormycosis. J Clin Microbiol 50:1491–1494. https://doi.org/10.1128/JCM.06611-11
Yamamoto K, Degawa Y, Yamada A (2020) Taxonomic study of Endogonaceae in the Japanese islands: new species of Endogone, Jimgerdemannia, and Vinositunica, gen. nov. Mycologia 112:309–328. https://doi.org/10.1080/00275514.2019.1689092
Yang LW, Ho HM, Chien CY (2012) Notes on Zygomycetes of Taiwan 10: the genus Lichtheimia in Taiwan. Fungal Sci 27:109–120
Yang Y, Liu XY, Huang B (2022) The complete mitochondrial genome of Linnemannia amoeboidea (W. Gams) Vandepol & Bonito (Mortierellales: Mortierellaceae). Mitochondrial DNA B 7:374–376. https://doi.org/10.1080/23802359.2022.2039080
Yao LD, Ju X, James TY et al (2018) Relationship between saccharifying capacity and isolation sources for strains of the Rhizopus arrhizus complex. Mycoscience 59:409–414. https://doi.org/10.1016/j.myc.2018.02.011
Yao YJ, Pegler DN, Young TW (1996) Genera of Endogonales. Royal Botanic Gardens, Kew
Yamamoto K, Degawa Y, Yamada A (2019) Taxonomic study of Endogonaceae in the Japanese islands: new species of Endogone, Jimgerdemannia, and Vinositunica, gen. nov. Mycologia 112:309–328. https://doi.org/10.1080/00275514.2019.1689092
Yin LQ, Zhang YZ, Azi F et al (2021) Neuroprotective potency of Tofu bio-processed using Actinomucor elegans against hypoxic injury induced by cobalt chloride in PC12 cells. Molecules 26:2983. https://doi.org/10.3390/MOLECULES26102983
Yip HY (1986) Umbelopsis fusiformis sp. nov. from a wet sclerophyll forest and a new combination for Mortierella ovata. Trans Br Mycol Soc 86:334–337. https://doi.org/10.1016/S0007-1536(86)80167-X
Young T (1987) Morphology of the anamorph of Hesseltinella vesiculosa. Trans Br Mycol Soc 89:392–396. https://doi.org/10.1016/S0007-1536(87)80126-2
Yuan HS, Lu X, Dai YC et al (2020) Fungal diversity notes 1277–1386: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 104:1–266. https://doi.org/10.1007/s13225-020-00461-7
Zhang ZY, Zhao YX, Shen X et al (2020) Molecular phylogeny and morphology of Cunninghamella guizhouensis sp. nov. (Cunninghamellaceae, Mucorales), from soil in Guizhou, China. Phytotaxa 455:31–39. https://doi.org/10.11646/phytotaxa.455.1.4
Zhao H, Lv ML, Liu Z et al (2021a) High-yield oleaginous fungi and high-value microbial lipid resources from Mucoromycota. BioEnerg Res 14:1196–1206. https://doi.org/10.1007/s12155-020-10219-3
Zhao H, Nie Y, Zong TK et al (2022) Three new species of Absidia (Mucoromycota) from China based on phylogeny, morphology and physiology. Diversity 14:132. https://doi.org/10.3390/d14020132
Zhao H, Nie Y, Zong TK et al (2022b) Species diversity and ecological habitat of Absidia (Cunninghamellaceae, Mucorales) with emphasis on five new species from forest and grassland soil in China. J Fungi 8:471. https://doi.org/10.3390/jof8050471
Zhao H, Nie Y, Jiang Y et al (2022) Comparative genomics of Mortierellaceae provides insights into lipid metabolism: two novel types of fatty acid synthase. J Fungi 8:891
Zhao H, Zhu J, Zong TK et al (2021b) Two new species in the family Cunninghamellaceae from China. Mycobiology 49:142–150. https://doi.org/10.1080/12298093.2021.1904555
Zhao RL, Li GJ, Sanchez-Ramirez S et al (2017) A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers 84:43–74. https://doi.org/10.1007/s13225-017-0381-5
Zhao Y, Wang H, Liu T et al (2014) The individual lipid compositions produced by Cunninghamella sp. Salicorn 5, an endophytic oleaginous fungus from Salicornia bigelovii Torr. Eur Food Res Tech 238:621–633. https://doi.org/10.1007/s00217-013-2141-4
Zheng HD, Zhuang WY, Wang XC et al (2020) Ascomycetes from the Qilian Mountains, China—Pezizomycetes and Leotiomycetes. Mycosystema 39:1823–1845. https://doi.org/10.13346/j.mycosystema.200217
Zheng XY, Chen JJ (2022) First report of leaf spot on pecan caused by Choanephora cucurbitarum in China. Plant Dis. https://doi.org/10.1094/PDIS-09-22-2240-PDN
Zheng RY, Chen GQ (1986) Blakeslea sinensis sp. nov., a further proof for retaining the genus Blakeslea. Acta Mycologica Sinica Suppl I:40–55
Zheng RY, Chen GQ (1991) A nonthermophilic Rhizomucor causing human primary cutaneous mucormycosis. Mycosystema 4:45–57
Zheng RY, Chen GQ (1998) Cunninghamella clavata sp. nov., a fungus with an unusual type of branching of sporophore. Mycotaxon 69:187–198
Zheng RY, Chen GQ (2001) A monograph of Cunninghamella. Mycotaxon 80:1–75
Zheng RY, Chen GQ, Hu FM (1988) Monosporus varieties of Syncephalastrum. Mycosystema 1:35–52
Zheng RY, Chen GQ, Huang H et al (2007) A monograph of Rhizopus. Sydowia 59:273
Zheng RY, Jiang HJM (1995) Rhizomucor endophyticus sp. nov., an endophytic zygomycetes from higher plants. Mycotaxon 56:455–466
Zheng RY, Liu XY (2005) Actinomucor elegans var. meitauzae, the correct name for A. taiwanensis and Mucor meitauzae (Mucorales, Zygomycota). Nova Hedwigia, pp 419–432
Zheng RY, Liu XY (2009) Taxa of Pilaira (Mucorales, Zygomycota) from China. Nova Hedwigia 88:255–267. https://doi.org/10.1127/0029-5035/2009/0088-0255
Zheng RY, Liu XY (2014) Ambomucor gen. & spp. nov. from China. Mycotaxon 126:97–108. https://doi.org/10.5248/126.97
Zheng RY, Liu XY, Li RY (2009) More Rhizomucor causing human mucormycosis from China: R. chlamydosporus sp. nov. Sydowia 61:135–147
Zheng RY, Liu XY, Wang YN (2013) Two taxa of the new record genus Backusella from China. Mycosystema 32:330–341
Zheng RY, Liu XY, Wang YN (2017) Circinella (Mucorales, Mucoromycotina) from China. Mycotaxon 132:43–62. https://doi.org/10.5248/132.43
Zheng RY (2002) Zygorhynchus multiplex, a new species isolated from paddy soil. Mycotaxon 84:367–378
Zimin AV, Marçais G, Puiu D et al (2013) The MaSuRCA genome assembler. Bioinformatics 29:2669–2677. https://doi.org/10.1093/bioinformatics/btt476
Zoghi M, Gandomkar S, Habibi Z (2019) Biotransformation of progesterone and testosterone enanthate by Circinella muscae. Steroids 151:108446. https://doi.org/10.1016/j.steroids.2019.108446
Zong TK, Zhao H, Liu XL et al (2021) Taxonomy and phylogeny of four new species in Absidia (Cunninghamellaceae, Mucorales) from China. Front Microbiol 12:677836. https://doi.org/10.3389/fmicb.2021.677836
Zycha H, Siepmann R, Linnemann G (1969) Mucorales: eine Beschreibung aller Gattungen und Arten dieser Pilzgruppe. Verlag von J. Cramer pp 1–355.
Acknowledgements
We thank the following scholars for their help with sampling and culture isolation: Yu-Chuan Bai and Peng-Cheng Deng (North Minzu University), Meng Zhou, Hong-Ming Zhou, Shun Liu, Xiao-Wu Man, Chang-Ge Song and Tai-Min Xu (Beijing Forestry University), Ru-Yong Zheng, He Huang, Hong-Mei Liu, Ya-Ning Wang, Jing Yang, Mao-Qiang He, Yue Zhang, Xing-Chun Li, Jie Li, Xiao-Ling Liu, Gui-Qing Chen, Feng-Yan Bai, Xue-Wei Wang (Prof.), Xue-Wei Wang (Ph.D. student), Xin-Yu Zhu and Long Wang (Institute of Microbiology, Chinese Academy of Chinese), Xiao Ju (Nanjing Agricultural University), Zhi-Kang Zhang (Ludong University), Ze Liu, Xiang Sun and Qi-Ming Wang (Hebei University), Zhong-Dong Yu (Northwest A&F University), Yan-Yan Long (Guangxi Academy of Agricultural Sciences), Nian-Kai Zeng (Hainan Medical University), Ze-Fen Yu and Min Qiao (Yunnan University), Pei-Wu Cui (Hunan University of Chinese Medicine), Jun-Feng Liang (Chinese Academy of Forestry), Yan-Feng Hang (Guizhou University), Wang-Qiu Deng (Guangdong Institute of Microbiology), Pu Liu, Li-Ying Ren (Jilin Agricultural University), Biao Xu (Tarim University), Rafael F. Castañeda-Ruiz (Academia de Ciencias de Cuba, Cuba), Kerstin Voigt (Friedrich Schiller University Jena, Germany), Manfred Binder (Clark University, USA), and Timothy Y. James (Michigan University, USA), Nicolas Corradi and Mathu Malar C (University of Ottawa, USA), and Francis Michel Martin (Head of Department at French National Institute for Agriculture, Food, and Environment, France).
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31970009, 32170012 and 32000010), the Second Tibetan Plateau Scientific Expedition and Research Program (STEP, Grant No. 2019QZKK0503), and the Third Xinjiang Scientific Expedition and Research Program (STEP, Grant No. 2021XJKK0505).
Author information
Authors and Affiliations
Contributions
Heng Zhao is responsible for culture isolations, identifications and descriptions, data analyses, drafting and editing the manuscript; Yu-Cheng Dai and Xiao-Yong Liu for funding acquisition; Yuan Yuan, Yu-Cheng Dai and Xiao-Yong Liu conceiving this study, and improving the manuscript; Yong Nie, Tong-Kai Zong, Ke Wang, Mei-Lin Lv, Yu-Jin Cui, Ablat·Tohtirjap, Jia-Jia Chen, Chang-Lin Zhao, Fang Wu and Bao-Kai Cui contributed to the sample collection and data analyses.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no conflict of interest.
Additional information
Handling Editor: Jian-Kui Liu.
Supplementary Information
Below is the link to the electronic supplementary material.
Fig. S1
. Species information concerning publication years, type’ origins, and first record time in China. (XLSX 33 kb)
Fig. S1
. A Maximum Likelihood phylogenetic tree illustrating relationships within subkingdom Mucoromyceta based on ITS and LSU rDNA sequences, with Neurospora crassa as outgroup. New taxa proposed in the present study are in red. Maximum Likelihood bootstrap values (≥50%) / Bayesian Inference (BI) Posterior Probabilities (≥0.80) are indicated along branches. Taxa are highlighted in different colors. A scale bar in the upper left indicates substitutions per site. (PDF 92 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, H., Nie, Y., Zong, TK. et al. Species diversity, updated classification and divergence times of the phylum Mucoromycota. Fungal Diversity 123, 49–157 (2023). https://doi.org/10.1007/s13225-023-00525-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-023-00525-4