Abstract
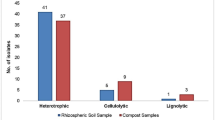
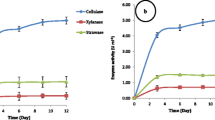
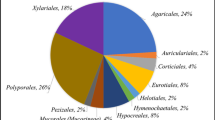
Lignocellulosic wastes such as straw are attractive resources for biofuel production when subjected to biological treatment (hydrolysis). However, their complex lignocellulosic structure can hinder saccharification. The urgent need for microbial groups with high levels of straw saccharifying activities is therefore a key step in the bioconversion of lignocellulosic straw into fermentable monosaccharides. Existing traditional methods of qualitative and quantitative screening of lignocellulolytic microbial isolates are costly, time consuming and largely not environmentally friendly. In this study, a Biolog (MT2) microplate-based assay was evaluated for potential use as an alternative screening method. This was carried out using three commercially available substrates (cellulose, xylan and lignin) and four native lignocellulosic straws (wheat, rice, sugarcane, and pea ball-milled straws). Selected bacterial isolates from soil, compost and straws were screened quantitatively using both traditional crude enzyme and Biolog (MT2) microplate methods. Positive correlations (R 2 values up to 0.86) between Biolog and the traditional enzyme methodologies were observed with respect to these isolates and their lignocellulosic activities. Quantitative assays were less labor intensive and faster (3–7 days) in Biolog microplates than in traditional assays which lasted for 12–15 days. Ball-milled rice and sugarcane straws were bio-converted to monosaccharaides more readily than wheat and pea straws and the commercially available substrates (cellulose, xylan and lignin). Environmental scanning electron microscopy (ESEM) analysis of ball-milled rice and sugarcane straws suggested that this was due to their higher silica content. Overall, the Biolog (MT2) microplate system was shown to be an effective, time saving and inexpensive alternative method for the screening of both lignocellulose-degrading bacteria and different substrates for saccharification.





Similar content being viewed by others
References
Adetutu EM, Thorpe K, Shahsavari E, Bourne S, Cao X, Mazaheri Nezhad Fard R, Kirby G, Ball AS (2012) Bacterial community survey of sediments at Naracoorte Caves, Australia. Int J Speleol 41(2):2
Ali MY, Rahman M, Rahman A, Basaglia M, Rahman M, Sultana T, Casella S (2014) Isolation of Bacillus spp. From soil and an evaluation of their sensitivity towards different extracts and essential oils of cumin (Cuminum cyminum L.). J Agric Sci Technol 16(3):623–633
Alvira P, Ballesteros M, Negro MJ (2013) Progress on enzymatic saccharification technologies for biofuels production. In: Gupta VK, Touhy M (eds) Biofuel technologies. Springer, Berlin, pp 145–169
Archana A, Satyanarayana T (1997) Xylanase production by thermophilic Bacillus licheniformis A99 in solid-state fermentation. Enzym Microb Technol 21(1):12–17
Bailey MJ, Biely P, Poutanen K (1992) Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol 23(3):257–270. doi:10.1016/0168-1656(92)90074-J
Ball AS, McCarthy AJ (1988) Saccharification of straw by actinomycete enzymes. J Gen Microbiol 134(8):2139–2147
Bandounas L, Wierckx N, de Winde J, Ruijssenaars H (2011) Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol 11(1):94
Banerjee S, Mudliar S, Sen R, Giri B, Satpute D, Chakrabarti T, Pandey R (2010) Commercializing lignocellulosic bioethanol: technology bottlenecks and possible remedies. Biofuels Bioprod Biorefin 4(1):77–93
Binod P, Sindhu R, Singhania RR, Vikram S, Devi L, Nagalakshmi S, Kurien N, Sukumaran RK, Pandey A (2010) Bioethanol production from rice straw: an overview. Bioresour Technol 101(13):4767–4774
Boonmee A (2009) Screening of rice straw degrading microorganisms and their cellulase activities. KKU Sci J 37:83–88
Brito-Cunha CCD, de Campos ITN, de Faria FP, Bataus LAM (2013) Screening and xylanase production by streptomyces sp grown on lignocellulosic wastes. Appl Biochem Biotechnol 170(3):598–608. doi:10.1007/s12010-013-0193-3
Bushnell LD, Haas HF (1941) The utilization of certain hydrocarbons by microorganisms. J Bacteriol 41(5):653–673
Chandrakant P, Bisaria VS (1998) Simultaneous bioconversion of cellulose and hemicellulose to ethanol. Crit Rev Biotechnol 18(4):295–331. doi:10.1080/0738-859891224185
Chou C-H, Han C-L, Chang J-J, Lay J-J (2011) Co-culture of Clostridium beijerinckii L9, Clostridium butyricum M1 and Bacillus thermoamylovorans B5 for converting yeast waste into hydrogen. Int J Hydrog Energy 36(21):13972–13983
Chun J, Bae KS (2000) Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek 78(2):123–127
Das H, Singh SK (2004) Useful byproducts from cellulosic wastes of agriculture and food industry: a critical appraisal. Crit Rev Food Sci Nutr 44(2):77–89
Dashtban M, Schraft H, Qin WS (2009) Fungal bioconversion of lignocellulosic residues; opportunities and perspectives. Int J Biol Sci 5(6):578–595
Dashtban M, Maki M, Leung KT, Mao C, Qin W (2010) Cellulase activities in biomass conversion: measurement methods and comparison. Crit Rev Biotechnol 30(4):302–309
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M (2008) Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36(suppl 2):W465–W469
Domínguez‐Escribá L, Porcar M (2010) Rice straw management: the big waste. Biofuels Bioprod Biorefin 4(2):154–159
dos Santos LF, Defrenne L, Krebs-Brown A (2002) Comparison of three microbial assay procedures for measuring toxicity of chemical compounds: ToxAlert (R) 10, cell sense and biolog MT2 microplates. Anal Chim Acta 456(1):41–54. doi:10.1016/S0003-2670(01)00907-2
Frias M, Villar-Cociña E, Valencia-Morales E (2007) Characterisation of sugar cane straw waste as pozzolanic material for construction: Calcining temperature and kinetic parameters. Waste Manag 27(4):533–538
Galbe M, Zacchi G (2007) Pretreatment of lignocellulosic materials for efficient bioethanol production. In: Olsson L (ed) Biofuels. Advances in Biochemical Engineering/Biotechnology, vol 8. Springer, Berlin, pp 41–65
Ghose TK (1987) Measurement of cellulase activities. Pure Appl Chem 59(2):257–268. doi:10.1351/pac198759020257
Gusakov AV (2011) Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol 29(9):419–425. doi:10.1016/j.tibtech.2011.04.004
Haruta S, Cui Z, Huang Z, Li M, Ishii M, Igarashi Y (2002) Construction of a stable microbial community with high cellulose-degradation ability. Appl Microbiol Biotechnol 59(4–5):529–534
Holtzapple M, Cognata M, Shu Y, Hendrickson C (1990) Inhibition of Trichoderma reesei cellulase by sugars and solvents. Biotechnol Bioeng 36(3):275–287
Howard R, Abotsi E, Van Rensburg EJ, Howard S (2004) Lignocellulose biotechnology: issues of bioconversion and enzyme production. Afr J Biotechnol 2(12):602–619
Huang C-H, Chang M-T, Huang L, Chu W-S (2012) Development of a novel PCR assay based on the gyrase B gene for species identification of Bacillus licheniformis. Mol Cell Probes 26(5):215–217
Ibrahim ASS, El-diwany AI (2007) Isolation and identification of new cellulases producing thermophilic bacteria from an Egyptian hot spring and some properties of the crude enzyme. Aust J Basic Appl Sci 1(4):473–478
Jørgensen H, Kristensen JB, Felby C (2007) Enzymatic conversion of lignocellulose into fermentable sugars: challenges and opportunities. Biofuels Bioprod Biorefin 1(2):119–134
Kadali KK, Simons KL, Skuza PP, Moore RB, Ball AS (2012) A complementary approach to identifying and assessing the remediation potential of hydrocarbonoclastic bacteria. J Microbiol Methods 88(3):348–355
Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A (2008) A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr Microbiol 57(5):503–507
Kumar P, Barrett DM, Delwiche MJ, Stroeve P (2009) Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res 48(8):3713–3729. doi:10.1021/Ie801542g
Lu WJ, Wang HT, Huang CY, Reichardt W (2008) Aromatic compound degradation by iron reducing bacteria isolated from irrigated tropical paddy soils. J Environ Sci China 20(12):1487–1493
Lucas F, Broennimann O, Febbraro I, Heeb P (2003) High diversity among feather-degrading bacteria from a dry meadow soil. Microb Ecol 45(3):282–290
Maki ML, Idrees A, Leung KT, Qin W (2012) Newly isolated and characterized bacteria with great application potential for decomposition of lignocellulosic biomass. J Mol Microbiol Biotechnol 22(3):156–166
Manage PM, Edwards C, Singh BK, Lawton LA (2009) Isolation and identification of novel microcystin-degrading bacteria. Appl Environ Microbiol 75(21):6924–6928. doi:10.1128/Aem. 01928-09
Michelin M, Maria de Lourdes T, Ruzene DS, Silva DP, Teixeira JA (2013) Application of lignocelulosic residues in the production of cellulase and hemicellulases from fungi. In: Polizeli M de Lourdes TM, Rai M (ed) Fungal enzymes. CRC, Boca Raton, pp 31–64
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428. doi:10.1021/Ac60147a030
Mosier N, Wyman C, Dale B, Elander R, Lee Y, Holtzapple M, Ladisch M (2005) Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96(6):673–686
Murad HA, Azzaz HEDH (2013) Cellulase production from rice straw by Aspergillus flavus NRRL 552. Sci Int 1(4)103–107
Nelson N (1944) A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem 153(2):375–379
Obruca S, Marova I, Matouskova P, Haronikova A, Lichnova A (2012) Production of lignocellulose-degrading enzymes employing Fusarium solani F-552. Folia Microbiol 57(3):221–227
Pandey S, Singh S, Yadav AN, Nain L, Saxena AK (2013) Phylogenetic diversity and characterization of novel and efficient cellulase producing bacterial isolates from various extreme environments. Biosci Biotechnol Biochem 77(7):1474–1480
Pérez J, Munoz-Dorado J, de la Rubia T, Martinez J (2002) Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int Microbiol 5(2):53–63
Plecha S, Hall D, Tiquia-Arashiro SM (2013) Screening for novel bacteria from the bioenergy feedstock switchgrass (Panicum virgatum L.). Environ Technol 34(13–14):1895–1904
Pohlschroeder M, Leschine SB, Canale-Parola E (1994) Spirochaeta caldaria sp. nov., a thermophilic bacterium that enhances cellulose degradation by Clostridium thermocellum. Arch Microbiol 161(1):17–24
Prasanna P, Patel M (1987) SEM studies on silica from plant materials. X-Ray Spectrom 16(2):57–60
Saha BC (2003) Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30(5):279–291. doi:10.1007/s10295-003-0049-x
Sanchez C (2009) Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol Adv 27(2):185–194. doi:10.1016/j.biotechadv.2008.11.001
Sewalt V, Glasser W, Beauchemin K (1997) Lignin impact on fiber degradation. 3. Reversal of inhibition of enzymatic hydrolysis by chemical modification of lignin and by additives. J Agric Food Chem 45(5):1823–1828
Singh P, Suman A, Tiwari P, Arya N, Gaur A, Shrivastava A (2008) Biological pretreatment of sugarcane trash for its conversion to fermentable sugars. World J Microbiol Biotechnol 24(5):667–673
Soni SK, Batra N, Bansal N, Soni R (2010) Bioconversion of sugarcane bagasse into second generation bioethanol after enzymatic hydrolysis with in-house produced cellulases from Aspergillus sp. S4B2F. BioResour 5(2):741–757
Taherdanak M, Zilouei H (2014) Improving biogas production from wheat plant using alkaline pretreatment. Fuel 115:714–719
Taherzadeh MJ, Karimi K (2008) Pretreatment of lignocellulosic wastes to improve ethanol and biogas production: a review. Int J Mol Sci 9(9):1621–1651
Tran D-T, Lin C-W (2013) Developing co-culture system of dominant cellulolytic Bacillus sp. THLA0409 and dominant ethanolic Klebsiella oxytoca THLC0409 for enhancing ethanol production from lignocellulosic materials. J Taiwan Inst Chem Eng 44(5):762–769
Tuomela M, Vikman M, Hatakka A, Itavaara M (2000) Biodegradation of lignin in a compost environment: a review. Bioresour Technol 72(2):169–183. doi:10.1016/S0960-8524(99)00104-2
Wang H, Guo P, Wang X, Wang X, Cui Z (2012) Degrading capability and activity of extracellular xylanase secreted by a composite microbial system XDC-2. Afr J Biotechnol 11(34):8476–8483
Wu B, Zhao Y, Gao PJ (2007) A new approach to measurement of saccharifying capacities of crude cellulase. BioResources 1(2):189–200
Wu Q, Lu Y-F, Shi J-Z, Liang S-X, Shi J-S, Liu J (2011) Chemical form of metals in traditional medicines underlines potential toxicity in cell cultures. J Ethnopharmacol 134(3):839–843
Zimmermann W (1990) Degradation of lignin by bacteria. J Biotechnol 13(2):119–130
Acknowledgments
The authors are grateful to RMIT University and the Egyptian Government for the provision of a PhD scholarship to Mohamed Taha. The authors also acknowledge the facilities, and the scientific and technical assistance, of the Australian Microscopy & Microanalysis Research Facility (AMMRF) at the RMIT Microscopy & Microanalysis Facility, at RMIT University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Enzyme activities (cellulase, xylanase and strawase U mL−1) of the 30 different bacterial isolates using the traditional crude enzyme production assay method (DOCX 949 kb)
Supplementary Figure 1
Phylogenetic tree of selected bacterial isolates. Note: tree was constructed from 1,000–1,200 nucleotide positions. Distances were calculated with the maximum likelihood model using PhyML. Bootstrap values > 0.50 are shown. Asterisks refer to lowest enzyme activity (based on strawase assay) by the traditional assays and to the “negative isolates” by Biolog (MT2) microplates.(DOCX 18 kb)
Rights and permissions
About this article
Cite this article
Taha, M., Kadali, K.K., AL-Hothaly, K. et al. An effective microplate method (Biolog MT2) for screening native lignocellulosic-straw-degrading bacteria. Ann Microbiol 65, 2053–2064 (2015). https://doi.org/10.1007/s13213-015-1044-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-015-1044-y