Abstract
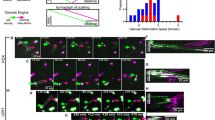
Enhanced motility in malignant cell is one hallmark of tumor metastasis. The clinical reality is that each of us responds differently to treatment, driving a significant interest in the development of therapies that are “right” to the individual. A biomedical analytical tool with low-cost, sensitive and short assaytime is critical for personalized medicine. Herein, a cost-effective and user-friendly microfluidic device was developed for studying of cell migration. A two-step photolithography procedure was conducted to reveal microchannels and microchambers with different depth. The PDMS/glass slide hydride device was assembled between a polymethyl methacrylate (PMMA) clamp which can adjust the pressure imposed on the device to control the fluid communication between mainchambers, thus identical wound (cell-free space) with clear edge can be easily formed within channel without extra chemical, mechanical force, fluidic manipulation and sophisticated microstructure. Using this device, we evaluated the combinatory of BRAFV600E inhibitor vemurafenib and epidermal growth factor receptor (EGFR) inhibitor gefitinib in inhibiting of melanoma cell migration with only 20 μL cell consumption, highlighting its potential in assaying rare clinical biopsy for personalized medicine. In addition, the on-chip migration model followed strictly follow the principle of conventional in vitro scratch/wound healing assay, facilitating it is translation to biologist.
Similar content being viewed by others
References
Lauffenburger, D.A. & Horwitz, A.F. Cell migration: a physically integrated molecular process. Cell 84, 359–369 (1996).
Wang, J., Petefish, J.W., Hillier, A.C. & Schneider, I.C. Epitaxially Grown Collagen Fibrils Reveal Diversity in Contact Guidance Behavior among Cancer Cells. Langmuir 31, 307–314 (2015).
Hou, Y., Rodriguez, L.L., Wang, J. & Schneider, I.C. Collagen attachment to the substrate controls cell clustering through migration. Phys. Biol. 11, 056007 (2014).
Eccles, S.A., Box, C. & Court, W. Cell migration/invasion assays and their application in cancer drug discovery. Biotechnol. Annu. Rev. 11, 391–421 (2005).
Liang, C.C., Park, A.Y. & Guan, J.L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2, 329–333 (2007).
Riahi, R., Yang, Y., Zhang, D.D. & Wong, P.K. Advances in wound-healing assays for probing collective cell migration. J. Lab. Autom. 17, 59–65 (2012).
Kunze, A., Pushkarsky, I., Kittur, H. & Di Carlo, D. Research highlights: measuring and manipulating cell migration. Lab Chip 14, 4117–4121 (2014).
Boneschansker, L., Yan, J., Wong, E., Briscoe, D.M. & Irimia, D. Microfluidic platform for the quantitative analysis of leukocyte migration signatures. Nat. Commun. 5, 4787 (2014).
Lamberti, G. et al. Bioinspired microfluidic assay for in vitro modeling of leukocyte-endothelium interactions. Anal. Chem. 86, 8344–8351 (2014).
Cheng, S.Y. et al. A hydrogel-based microfluidic device for the studies of directed cell migration. Lab Chip 7, 763–769 (2007).
Nie, F.Q. et al. On-chip cell migration assay using microfluidic channels. Biomaterials 28, 4017–4022 (2007).
van der Meer, A.D., Vermeul, K., Poot, A.A., Feijen, J. & Vermes, I. A microfluidic wound-healing assay for quantifying endothelial cell migration. Am. J. Physiol. Heart Circ. Physiol. 298, H719–725 (2010).
Jowhar, D., Wright, G., Samson, P.C., Wikswo, J.P. & Janetopoulos, C. Open access microfluidic device for the study of cell migration during chemotaxis. Integr. Biol. (Camb) 2, 648–658 (2010).
Zervantonakis, I.K. et al. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. USA 109, 13515–13520 (2012).
Shin, M.K., Kim, S.K. & Jung, H. Integration of intraand extravasation in one cell-based microfluidic chip for the study of cancer metastasis. Lab Chip 11, 3880–3887 (2011).
Jeong, G.S. et al. Microfluidic assay of endothelial cell migration in 3D interpenetrating polymer semi-network HA-Collagen hydrogel. Biomed. Microdevices 13, 717–723 (2011).
Chaw, K.C., Manimaran, M., Tay, E.H. & Swaminathan, S. Multi-step microfluidic device for studying cancer metastasis. Lab Chip 7, 1041–1047 (2007).
Chaw, K.C., Manimaran, M., Tay, F.E. & Swaminathan, S. Matrigel coated polydimethylsiloxane based microfluidic devices for studying metastatic and non-metastatic cancer cell invasion and migration. Biomed. Microdevices 9, 597–602 (2007).
Wu, J., Wu, X. & Lin, F. Recent developments in microfluidics-based chemotaxis studies. Lab Chip 13, 2484–2499 (2013).
Zhang, M., Li, H., Ma, H. & Qin, J. A simple microfluidic strategy for cell migration assay in an in vitro wound-healing model. Wound Repair Regen. 21, 897–903 (2013).
Zheng, C. et al. Live cell imaging analysis of the epigenetic regulation of the human endothelial cell migration at single-cell resolution. Lab Chip 12, 3063–3072 (2012).
Yu, L. et al. The CSPG4-specific monoclonal antibody enhances and prolongs the effects of the BRAF inhibitor in melanoma cells. Immunol. Res. 50, 294–302 (2011).
Nazarian, R. et al. Melanomas acquire resistance to B-RAF (V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010).
Flaherty, K.T. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010).
Chapman, P.B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. McArthur, N. Engl. J. Med. 364, 2507–2516 (2011).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF (V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Patel, S.P. et al. A phase II study of gefitinib in patients with metastatic melanoma. Melanoma Res. 21, 357–363 (2011).
Djerf, E.A. et al. ErbB receptor tyrosine kinases contribute to proliferation of malignant melanoma cells: inhibition by gefitinib (ZD1839). Melanoma Res. 19, 156–166 (2009).
Girotti, M.R. et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discovery 3, 158–167 (2013).
Gross, A. et al. Expression and activity of EGFR in human cutaneous melanoma cell lines and influence of vemurafenib on the EGFR pathway. Target. Oncol. 10, 77–84 (2015).
Sun, C. et al. Reversible and adaptive resistance to BRAF (V600E) inhibition in melanoma. Nature 508, 118–122 (2014).
Ong, F.S. et al. Personalized medicine and pharmacogenetic biomarkers: progress in molecular oncology testing. Expert Rev. Mol. Diagn. 12, 593–602 (2012).
Jackson, S.E. & Chester, J.D. Personalised cancer medicine. Int. J. Cancer 201, 137, 262–6 (2015)
Joseph, E.W. et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc. Natl. Acad. Sci. USA 107, 14903–14908 (2010).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gao, A., Tian, Y., Shi, Z. et al. A cost-effective microdevice bridges microfluidic and conventional in vitro scratch / wound-healing assay for personalized therapy validation. BioChip J 10, 56–64 (2016). https://doi.org/10.1007/s13206-016-0108-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-016-0108-9