Abstract
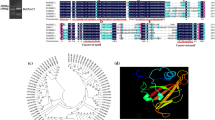
Plant nuclear factor (NF-Y) is a transcription activating factor, consisting of three subunits, and plays a key regulatory role in many stress-responsive mechanisms including drought and salinity stresses. NF-Ys function both as complex and individual subunits. Considering the importance of sugarcane as a commercial crop with high socio-economic importance and the crop being affected mostly by water deficit stress and salinity stress causing significant yield loss, nuclear transcriptional factor NF-YB2 was focused in this study. Plant nuclear factor subunit B2 from Erianthus arundinaceus (EaNF-YB2), a wild relative of sugarcane which is known for its drought and salinity stress tolerance, and commercial Saccharum hybrid Co 86032 (ShNF-YB2) was isolated and characterized. Both EaNF-YB2 and ShNF-YB2 genes are 543 bp long that encodes for a polypeptide of 180 amino acid residues. Comparison of EaNF-YB2 and ShNF-YB2 gene sequences revealed nucleotide substitutions at nine positions corresponding to three synonymous and six nonsynonymous amino acid substitutions that resulted in variations in physiochemical properties. However, multiple sequence alignment (MSA) of NF-YB2 proteins showed conservation of functionally important amino acid residues. In silico analysis revealed NF-YB2 to be a hydrophilic and intracellular protein, and EaNF-YB2 is thermally more stable than that of ShNF-YB2. Phylogenetic analysis suggested the lower rate of evolution of NF-YB2. Subcellular localization in sugarcane callus revealed NF-YB2 localization at nucleus that further evidenced it to be a transcription activation factor. Comparative RT-qPCR experiments showed a significantly higher level of NF-YB2 expression in E. arundinaceus when compared to that in the commercial Saccharum hybrid Co 86032 under drought and salinity stresses. Hence, EaNF-YB2 could be an ideal candidate gene, and its overexpression in sugarcane through genetic engineering approach might enhance tolerance to drought and salinity stresses.








Similar content being viewed by others
Data availability statement
All relevant data are within the paper and its supporting information files.
References
Amalraj VA, Balasundaram N (2006) On the taxonomy of the members of ‘Saccharum complex’. Genet Resour Crop Evol 53:35–41
Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1:213–222
Anunanthini P, Manoj VM, Sarath Padmanabhan TS, Dhivya S, Ashwin Narayan J, Appunu C, Sathishkumar R (2019) In silico characterization and functional validation of membrane-bound cell signaling COLD1 gene in monocots during abiotic stress. Funct Plant Biol 46:524–532
Ashwin Narayan J, Dharshini S, Manoj VM, Sarath Padmanabhan TS, Kadirvelu K, Suresha GS, Subramonian N, Ram B, Premachandran MN, Appunu C (2019) Isolation and characterization of water deficit stress responsive α-expansin 1 (EXPA1) gene from Saccharum complex. 3 Biotech 9:186
Asif MA, Zafar Y, Iqbal J, Iqbal MM, Rashid U, Ali GM, Arif A, Nazir F (2011) Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea L.) improves salt and drought tolerence. Mol Biotechnol 49(3):250–256
Augustine SM, Narayan JA, Syamaladevi DP, Appunu C, Chakravarthi M, Ravichandran V, Subramonian N (2015a) Erianthus arundinaceus HSP70 (EaHSP70) overexpression increases drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Sci 232:23–34
Augustine SM, Syamaladevi DP, Premachandran MN, Ravichandran V, Subramonian N (2015b) Physiological and molecular insights to drought responsiveness in Erianthus spp. Sugar Tech 17:121–129
Augustine SM, Ashwin Narayan J, Divya PS, Appunu C, Chakravarthi M, Ravichandran V, Tuteja N, Subramonian N (2015c) Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Cell Rep 34:247–263
Ballif J, Endo S, Kotani M, MacAdam J, Wu Y (2011) Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol Biochem 49:579–583
Basnayake J, Jackson PAN, Inman-Bamber G, Lakshmanan P (2012) Sugarcane for water-limited environments. Genetic variation in cane yield and sugar content in response to water stress. J Exp Bot 63:6023–6033
Bentley GA, Lewit-Bentley A, Finch JT, Podjarny AD, Roth M (1984) Crystal structure of the nucleosome core particle at 16 Å resolution. J Mol Biol 176:55–75
Buxbaum E (2007) Fundamentals of protein structure and function. Springer, New York
Cai Q, Aitken K, Deng H, Chen X, Fu C, Jackson P, McIntyre C (2005) Verification of the introgression of Erianthus arundinaceus germplasm into sugarcane using molecular markers. Plant Breed 124(4):322–328
Cedano J, Aloy P, Perez-Pons JA, Querol E (1997) Relation between amino acid composition and cellular location of proteins. J Mol Biol 266:594–600
Chen M, Zhao Y, Zhuo C, Lu S, Guo Z (2015) Overexpression of a NF-YC transcription factor from Bermuda grass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol J 13:482–491
Chomzynski P, Mackey K (1995) Modification of the TRI reagent [TM] procedure for isolation of RNA from polysaccharide and proteoglycan-rich sources. Biotechiques 19:942–945
Chou KC (2004) Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21:10–19
Chou KC, Shen HB (2007) Large-scale plant protein subcellular location prediction. J Cell Biochem 100:665–678
Chou KC, Shen HB (2008) Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protocols 3:153–162
Chou KC, Shen HB (2010) Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5(6):e11335
D’Hont A, Glaszmann JC (2001) Sugarcane genome analysis with molecular markers, a first decade of research. Proc Int Soc Sugarcane Technol 24:556–559
D’Hont A, Rao PS, Feldmann P, Grivet L, Islam Faridi N, Taylor P, Glaszmann JC (1995) Identification and characterization of sugarcane intergeneric hybrids, Saccharum officinarum x Erianthus arundinaceus, with molecular markers and DNA in situ hybridization. Theor Appl Genet 91:320–326
Daniels J, Roach BT (1987) Taxonomy and evolution. In: Heinz A (ed) Sugarcane improvement through breeding. Elsevier, Amsterdam, pp 7–84
Deng Q, Dou Z, Chen J, Wang K, Shen W (2019) Drought tolerance evaluation of intergeneric hybrids of BC3F1 lines of Saccharum officinarum × Erianthus arundinaceus. Euphytica 215:207
D'Hont A, Grivet L, Feldmann P, Glaszmann JC, Rao S, Berding N (1996) Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp) by molecular cytogenetics. Mol Gen Genet MGG 250(4):405–413
Dolfini D, Gatta R, Mantovani R (2012) NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol 47:29–49
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Feng ZJ, He GH, Zheng WJ, Lu PP, Chen M, Gong YM, Ma YZ, Xu ZS (2015) Foxtail millet NF-Y families: genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front Plant Sci 6:1142
Garsmeur O, Droc G, Antonise R, Grimwood J, Potier B, Aitken K, Jenkins J, Martin G, Charron C, Hervouet C, Costet L (2018) A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat Comm 9:2638
Gentile A, Dias LI, Mattos RS, Ferreira TH, Menossi M (2015) MicroRNAs and drought responses in sugarcane. Front Plant Sci 6:58
Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182:319–326
Gnesutta N, Saad D, Chaves-Sanjuan A, Mantovani R, Nardini M (2017) Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the LEC1 family of NF-Y subunits in plants. Mol Plant 10:645–648
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2011) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40(D1):D1178–D1186
Guruprasad K, Reddy BB, Pandit MW (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng Design and Sel 4:155–161
Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, Grimm B (2012) Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol Plant 5:876–888
Han X, Tang S, An Y, Zheng DC, Xia XL, Yin WL (2013) Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J Exp Bot 64:4589–4601
Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H (2014) Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun 5:4601
Ikai A (1980) Thermostability and aliphatic index of globular proteins. J Biochem 88:1895–1898
Jackson P, Henry RJ (2011) Erianthus. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, industrial crops. Springer, Berlin, pp 97–107
Jin J, Zhang H, Kong L, Gao G, Luo J (2013) Plant TFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42(D1):D1182–D1187
Kahle J, Baake M, Doenecke D, Albig W (2005) Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin β and importin 13. Mol Cell Biol 25:5339–5354
Kim IS, Sinha S, De Crombrugghe B, Maity SN (1996) Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol Cell Biol 16:4003–4013
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Leyva-Gonzalez MA, Ibarra-Laclette E, Cruz-Ramirez A, Herrera-Estrella L (2012) Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7:e48138
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post transcriptionally to promote drought resistance. Plant Cell 20:2238–2251
Li C, Distelfeld A, Comis A, Dubcovsky J (2011) Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J 67:763–773
Li W, Mallano AI, Bo L, Wang T, Nisa Z, Li Y (2016) Soybean transcription factor GmNFYB1 confers abiotic stress tolerance to transgenic Arabidopsis plants. Can J Plant Sci 97:501–515
Liu JX, Howell SH (2010) bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22:782–796
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nat 389:251–260
Manoj VM, Anunanthini P, Swathik Clarancia P, Dharshini S, Ashwin Narayan J, Manickavasagam M, Sathishkumar R, Sursha GS, Hemaprabha G, Ram B, Appunu C (2019) Comparative functional analysis of Glyoxalase pathway genes in Erianthus arundinaceus and commercial sugarcane hybrid under salinity and drought conditions. BMC Genom 19:986
Mantovani R (1999) The molecular biology of the CCAAT-binding factor NF-Y. Gene 239:15–27
Maruyama KY, Todaka DA, Mizoi JU, Yoshida TA, Kidokoro SA, Matsukura SA, Takasaki HI, Sakurai TE, Yamamoto YY, Yoshiwara KY, Kojima MI (2011) Identification of cis-acting promoter elements in cold-and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res 19:37–49
Matsuo K, Chuenpreecha T, Ponragdee W, Wonwivachai C (2001) Eco-physiological characteristics and yielding ability of Erianthus spp. JIRCAS Res Highlights 12–13
Mcnabb DS, Tseng KA, Guarente L (1997) The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol Cell Biol 17:7008–7018
Nair NV, Mohanraj K, Sundaravelpandian K, Selvi A, Suganya A, Appunu C (2017) Characterization of an intergeneric hybrid of Erianthus procerus × Saccharum officinarum and its backcross progenies. Euphytica 213:267–277
Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5:170
Nardone V, Chaves-Sanjuan A, Nardini M (2017) Structural determinants for NF-Y/DNA interaction at the CCAAT box. Biochim Biophys Acta (BBA) Gene Regul Mech 1860:571–580
Palmeros-Suarez PA, Massange-Sánchez JA, Martinez-Gallardo NA, Montero-Vargas JM, Gomez-Leyva JF, Delano-Frier JP (2015) The overexpression of an Amaranthus hypochondriacus NF-YC gene modifies growth and confers water deficit stress resistance in Arabidopsis. Plant Sci 240:25–40
Piperidis G, Christopher MJ, Carroll BJ, Berding N, D’Hont A (2000) Molecular contribution to selection of intergeneric hybrids between sugarcane and the wild species Erianthus arundinaceus. Genome 43(6):1033–1037
Piperidis G, Piperidis N, D’Hont A (2010) Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genom 284:65–73
Premachandran MN, Sobhakumari VP, Lekshmi M, Raffee Viola V (2017) Genome characterization of in vitro induced amphiploids of an intergeneric hybrid Erianthus arundinaceus × Saccharum spontaneum. Sugar Tech 19:386–393
Quach TN, Nguyen HT, Valliyodan B, Joshi T, Xu D, Nguyen HT (2015) Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol Genet Genom 290:1095–1115
Ram B, Sahi BK, Sreenivasan TV, Singh N (2001) Introgression of low temperature tolerance and red rot resistance from Erianthus in sugarcane. Euphytica 122:145–153
Ramesh P (2000) Effect of different levels of drought during the formative phase on growth parameters and its relationship with dry matter accumulation in sugarcane. J Agron Crop Sci 185:83–89
Robertson MJ, Inmam-Bamber NG, Muchow RC, Wood AW (1999) Physiology and productivity of sugarcane with early and mid-season water deficit. Field Crop Res 64:211–227
Romier C, Cocchiarella F, Mantovani R, Moras D (2003) The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem 278:1336–1345
Sardana HR (2002) Evaluation of Saccharum barberi, S. sinense and Erianthus spp. clones of sugarcane for susceptibility to major borer pests. Ann Plant Prot Sci 10:42–44
Shen HB, Chou KC (2006) Ensemble classifier for protein fold pattern recognition. Bioinformatics 22:1717–1722
Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149:625–641
Sinha S, Kim IS, Sohn KY, De Crombrugghe B, Maity SN (1996) Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol Cell Biol 16:328–337
Srivasta A, Mehta S, Lindlof A, Bhargava S (2010) Over-represented promoter motifs in abiotic stress-induced DREB genes of rice and sorghum and their probable role in regulation of gene expression. Plant Signal Behav 5:775–784
Stephenson TJ, McIntyre CL, Collet C, Xue GP (2007) Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol 65:77–92
Sun X, Ren Y, Zhang X, Lian H, Zhou S, Liu S (2016) Overexpression of a garlic nuclear factor Y (NF-Y) B gene, AsNF-YB3, affects seed germination and plant growth in transgenic tobacco. Plant Cell Tiss Org Cult 127:513–523
Suprasanna P, Patade VY, Desai NS, Devarumath RM, Kawar PG, Pagariya MC, Ganapathi A, Manickavasagam M, Babu KH (2011) Biotechnological developments in sugarcane improvement: an overview. Sugar Tech 13(4):322–335
Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N (2008) Identification, characterization and interaction of HAP family genes in rice. Mol Genet Genom 279:279–289
Valarmathi R, Mahadeva Swamy HK, Appunu C (2018) Sugarcane root phenotyping for drought tolerance using customized PVC platforms. SBI NEWS 38(2):1–3
Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984
Xuanyuan G, Lu C, Zhang R, Jiang J (2017) Overexpression of StNF-YB3.1 reduces photosynthetic capacity and tuber production, and promotes ABA-mediated stomatal closure in potato (Solanum tuberosum L.). Plant Sci 261:50–59
Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58:843–856
Yang J, Wan XL, Guo C, Zhang JW, Bao MZ (2016) Identification and expression analysis of nuclear factor Y families in Prunus mume under different abiotic stresses. Biol Plant 60:419–426
Zhang T, Zhang D, Liu Y, Luo C, Zhou Y, Zhang L (2015) Overexpression of a NF-YB3 transcription factor from Picea wilsonii confers tolerance to salinity and drought stress in transformed Arabidopsis thaliana. Plant Physiol Biochem 94:153–164
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170
Acknowledgements
The authors would like to thank the Department of Biotechnology (Grant No. 102/IFD/SAN/1151/2017-2018), New Delhi, Government of India for funding support. We thank the Director, Centre for Plant Molecular Biology and Bioinformatics (CPMB&B) and the Head, Department of Bioinformatics, Tamil Nadu Agricultural University (TNAU), Coimbatore for extending Bioinformatics facility.
Author information
Authors and Affiliations
Contributions
SCP: conceptualization, design, methodology, performed the experiments and manuscript drafting. NM: vector construction. MVM, realtime PCR; SPTS, DS, ANJ, MC, VR: writing-review and editing. SGS: localization experiment. HG, BR: review and editing of the manuscript. AC: conceptualization, designed, performed the experiments, supervision, review and editing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Peter, S.C., Murugan, N., Mohanan, M.V. et al. Isolation, characterization and expression analysis of stress responsive plant nuclear transcriptional factor subunit (NF-YB2) from commercial Saccharum hybrid and wild relative Erianthus arundinaceus. 3 Biotech 10, 304 (2020). https://doi.org/10.1007/s13205-020-02295-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02295-1