Abstract
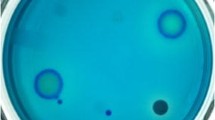
Here, we report on the isolation of bacterial isolates from Himalayan niches, which produced extracellular l-asparaginase with low/no glutaminase activity. From the 235 isolates, 85 asparaginase positive bacterial isolates were identified by qualitative screening using optimized chromogenic dyes assay. Optimized concentration of different dyes revealed maximum color visualization in phenol red (0.003%). The diversity analysis of asparaginase positive isolates revealed that Proteobacteria (83%) are the most dominant, followed by Actinobacteria (12%), Firmicutes (3%), and Bacteriodetes (2%). Eleven isolates, which represented seven Pseudomonas species, one species each of the genus Arthrobacter, Janthinobacterium, Lelliottia, and Rahnella, were selected for further studies based on highest zone ratio and novel aspects for l-asparaginase production. Of these, five isolates, namely, Pseudomonas sp. PCH133, Pseudomonas sp. PCH146, Pseudomonas sp. PCH182, Rahnella sp. PCH162, and Arthrobacter sp. PCH138, produced l-asparaginase without glutaminase activity after 55 h of growth with the former isolate showing the highest l-asparaginase activity (1.67 U/ml). Interestingly, this is the first report of l-asparaginase production by members of the genera Janthinobacterium, Rahnella, and Lelliottia.


Similar content being viewed by others
References
Alrumman SA, Mostafa YS, Kholood A, Al-izran Alfaifi MY, Taha TH, Elbehair SE (2019) Production and anticancer activity of an l-asparaginase from Bacillus licheniformis isolated from the Red Sea, Saudi Arabia. Sci Rep. 9:3756. https://doi.org/10.1038/s41598-019-40512-x
Andrade FA, Borges SK, Silveira VS (2014) Update on the use of l-asparaginase in infants and adolescent patients with acute lymphoblastic leukemia. Clin Med Insights Oncol 8:95–100. https://doi.org/10.4137/CMO.S10242
Badoei-Dalfard A (2016) l-asparaginase production in the Pseudomonas pseudoalcaligenes strain JHS-71 isolated from Jooshan Hot-spring. Mol Biol Res Commun 5:1–10. https://doi.org/10.22099/MBRC.2016.3379
Bansal S, Gnaneswari D, Mishra P, Kundu B (2010) Structural stability and functional analysis of l-asparaginase from Pyrococcus furiosus. Biochem (Moscow) 75:375–381. https://doi.org/10.1134/S0006297910030144
Batool T, Makky EA, Jalal M, Yusoff MM (2016) A comprehensive review on l-asparaginase and its applications. Appl Biochem Biotechnol 178:900–923. https://doi.org/10.1007/s12010-015-1917-3
Broome J (1963) Evidence that the l-asparaginase of guinea pig serum is responsible for its anti-lymphoma effects. J Exp Med 118:121–148. https://doi.org/10.1007/s12010-015-1917-310.1084/jem.118.1.99
Cappelletti D, Chiarelli LR, Pasquetto MV et al (2008) Helicobacter pylori l-asparaginase: a promising chemotherapeutic agent. Biochem Biophys Res Commun 377:1222–1226. https://doi.org/10.1016/j.bbrc.2008.10.118
Ciccazzo S, Esposito A, Rolli E, Zerbe S, Daffonchio D, Brusetti L (2014) Safe-site effects on rhizosphere bacterial communities in a high-altitude alpine environment. Biomed Res Int 2014:1–9. https://doi.org/10.1155/2014/480170
Davidson L, Burkom M, Ahn S, Chang LC, Kitto B (1977) l-asparaginases from Citrobacter freundii. Biochim Biophys Acta 480:282–294. https://doi.org/10.1016/0005-2744(77)90341-2
Dolowy WC, Henson D, Cornet J, Sellin H (1966) Toxic and antineoplastic effects of l-asparaginase: study of mice with lymphoma and normal monkeys and report on a child with leukemia. Cancer 19:1813–1819. https://doi.org/10.1002/1097-0142(196612)19:12%3c1813:AID-CNCR10%3e3.0.CO;2-E
Einsfeldt K, Baptista IC, Castanheira JC et al (2016) Recombinant l-asparaginase from Zymomonas mobilis: a potential new antileukemic agent produced in Escherichia coli. PLoS ONE 11(6):e0156692. https://doi.org/10.1371/journal.pone.0156692
El-Bessoumy AA, Sarhan M, Mansour J (2004) Production, isolation, and purification of l-asparaginase from Pseudomonas aeruginosa 50071 using solid-state fermentation. BMB Rep 37:387–393. https://doi.org/10.5483/BMBRep.2004.37.4.387
El-Nagga NE, El-Ewasy SM, El-Shweihy NM (2014) Microbial l-asparaginase as a potential therapeutic agent for the treatment of acute lymphoblastic leukemia: the pros and cons. Int J Pharmacol 10:182–199. https://doi.org/10.1007/s12010-015-1917-310.3923/ijp.2014.182.199
Erva RR, Goswami AN, Suman P, Vedanabhatla R, Rajulapati SB (2017) Optimization of l-asparaginase production from novel Enterobacter sp., by submerged fermentation using response surface methodology. Prep Biochem Biotechnol 16:219–228. https://doi.org/10.1080/10826068.2016.1201683
Feng Y, Liu S, Jiao Y, Gao H, Wang M, Du G, Chen J (2017) Enhanced extracellular production of l-asparaginase from Bacillus subtilis 168 by B. subtilis WB600 through a combined strategy. Appl Microbiol Biotechnol 101:1509–1520. https://doi.org/10.1007/s12010-015-1917-310.1007/s00253-016-7816-x
Ferbiyanto A, Rusmana I, Raffiudin R (2015) Characterization and identification of cellulolytic bacteria from gut of worker Macrotermes gilvus. HAYATI J Biosci 22:197–200. https://doi.org/10.1016/j.hjb.2015.07.001
Ghasemi A, Asad S, Kabiri M, Dabirmanesh B (2017) Cloning and characterization of Halomonas elongate l-asparaginase, a promising chemotherapeutic agent. Appl Microbiol Biotechnol 101(19):7227–7238. https://doi.org/10.1007/s00253-017-8456-5
Gulati R, Saxena RK, Gupta R (1997) A rapid plate assay for screening l-asparaginase producing micro-organisms. Lett Appl Microbiol 24:23–26. https://doi.org/10.1046/j.1472-765X.1997.00331.x
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
Husain I, Sharma A, Kumar S, Malik F (2016) Purification and characterization of glutaminase free asparaginase from Enterobacter cloacae: in vitro evaluation of cytotoxic potential against human myeloid leukemia HL-60 cells. PLoS One 11:1–27. https://doi.org/10.1371/journal.pone.0148877
Imada A, Igarasi S, Nakahama K, Isono M (1973) Asparaginase and glutaminase activities of microorganisms. J Gen Microbiol 76:85–99. https://doi.org/10.1099/00221287-76-1-85
Kidd JG (1953) Regression of transplanted lymphomas induced in vivo by means of normal guinea pig serum. Course of transplanted cancers of various kinds in mice and rats given guinea pig serum, horse serum, or rabbit serum. J Exp Med 98:565–582. https://doi.org/10.1084/jem.98.6.565
Killander D, Dohlwitz A, Engstedt L et al (1976) Hypersensitive reactions and antibody formation during l-asparaginase treatment of children and adults with acute leukemia. Cancer 37:220–228. https://doi.org/10.1002/1097-0142(197601)37:1%3c220:AID-CNCR23%3e3.0.CO;2-W
Krishnapura PR, Belur PD, Subramanya S (2016) A critical review on properties and applications of microbial l-asparaginases. Crit Rev Microbiol 42:720–737. https://doi.org/10.3109/1040841X.2015.1022505
Kumar S, Venkata DV, Pakshirajan K (2011) Purification and characterization of glutaminase-free l-Asparaginase from Pectobacterium carotovorum MTCC 1428. Bioresour Technol 102:2077–2082. https://doi.org/10.1016/j.biortech.2010.07.114
Kumar V, Thakur V, Ambika Kumar S, Singh D (2018) Bioplastic reservoir of diverse bacterial communities revealed along altitude gradient of Pangi-Chamba trans-Himalayan region. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fny144
Kuwabara T, Asep A, Prihanto Wakayama M, Takagi K (2015) Purification and characterization of Pseudomonas aeruginosa PAO1 asparaginase. Procedia Environ Sci 28:72–77. https://doi.org/10.1016/j.proenv.2015.07.011
Mahajan RV, Saran S, Saxena RK, Srivastava AK (2013) A rapid, efficient and sensitive plate assay for detection and screening of l-asparaginase-producing microorganisms. FEMS Microbiol Lett 341:122–126. https://doi.org/10.1111/1574-6968.12100
Mahajan RV, Kumar V, Rajendran V, Saran S, Ghosh PC, Saxena RK (2014) Purification and characterization of a novel and robust l-asparaginase having low-glutaminase activity from Bacillus licheniformis: in vitro evaluation of anti-cancerous properties. PLoS One. https://doi.org/10.1371/journal.pone.0099037
Meena B, Anburajan L, Sathish T, Vijaya Raghavan R, Dharani G, Vinithkumar NV, Kirubagaran R (2015) l-Asparaginase from Streptomyces griseus NIOT-VKMA29: optimization of process variables using factorial designs and molecular characterization of l-asparaginase gene. Sci Rep 5:12404. https://doi.org/10.1038/srep12404
Mihooliya NK, Nandal J, Swami L, Verma H, Chopra L, Sahoo DK (2017) A new pH indicator dye-based method for rapid and efficient screening of l-asparaginase producing microorganisms. Enz Microb Technol 107:72–81. https://doi.org/10.1016/j.enzmictec.2017.08.004
Morris EK, Caruso T, Buscot F (2014) Choosing and using diversity indices: insights for ecological applications from the German biodiversity exploratories. Ecol Evol 4:3514–3524. https://doi.org/10.1002/ece3.1155
Munaganti RK, Muvva V, Indupalli MD (2015) Studies on optimization of l-Asparaginase production by Arthrobacter kerguelensis VL-RK_09 isolated from Mango orchards. Int J Pharm Pharm Sci 7:112–115
Nguyen HA, Su Y, Lavie A (2016) Design and characterization of Erwinia chrysanthemi l-asparaginase variants with diminished l-glutaminase activity. J Biol Chem 291:17664–17676. https://doi.org/10.1074/jbc.M116.728485
Pritsa AA, Kyriakidis DA (2001) l-Asparaginase of Thermus thermophilus: purification properties and identification of essential amino acids for its catalytic activity. Mol Cell Biochem 216:93–101. https://doi.org/10.1023/A:1011066129771
Radha R, Arumugam N, Sathyanarayana Gummadi N (2018) Glutaminase free l-asparaginase from Vibrio cholerae: heterologous expression, purification and biochemical characterization. Int J Biol Macromol 111:129–138. https://doi.org/10.1016/j.ijbiomac.2017.12.165
Ramakrishnan MS, Joseph R (1996) Characterization of an extracellular asparaginase of Rhodosporidium toruloides CBS14 exhibiting unique physicochemical properties. Can J Microbiol 42:316–325. https://doi.org/10.1139/m96-047
Saxena A, Upadhyay R, Kango N (2015) Isolation and identification of actinomycetes for production of novel extracellular glutaminase free l-asparaginase. Ind J Exp Biol 53:786–793
Sindhu R, Manonmani HK (2018) Expression and characterization of recombinant l-asparaginase from Pseudomonas fluorescens. Protein Expr Purif 143:83–91. https://doi.org/10.1016/j.pep.2017.09.009
Stres B, Sul WJ, Murovec B, Tiedje JM (2013) Recently deglaciated high-altitude soils of the Himalaya: diverse environments, heterogeneous bacterial communities and long-range dust inputs from the upper troposphere. PLoS One 8:e76440. https://doi.org/10.1371/journal.pone.0076440
Thakur V, Kumar V, Kumar S, Singh D (2018) Diverse culturable bacterial communities with cellulolytic potential revealed from pristine habitat in Indian trans-Himalaya. Can J Microbiol 64:798–808. https://doi.org/10.1139/cjm-2017-0754
van den Berg H (2011) Asparaginase revisited. Leuk Lymphoma 52:168–178. https://doi.org/10.3109/10428194.2010.537796
Vimal A, Kumar A (2017) In vitro screening and in silico validation revealed key microbes for higher production of significant therapeutic enzyme l-asparaginase. Enz Microb Technol 98:9–17. https://doi.org/10.1016/j.enzmictec.2016.12.001
Vrooman LM, Supko JG, Neuberg DS et al (2010) Erwinia asparaginase after allergy to E. coli asparaginase in children with acute lymphoblastic leukemia. Pediatr Blood Cancer 54:199–205. https://doi.org/10.1002/pbc.22225
Warrell RP, Arlin ZA, Gee TS (1982) Clinical evaluation of succinylated Acinetobacter glutaminase-asparaginase in adult leukemia. Cancer Treat Rep 66:1479–1485
Acknowledgements
Authors duly acknowledge the technical assistance provided by Anil Kumar Chaudhary for 16S rDNA sequencing. This manuscript represents CSIR-IHBT Communication No. 4293.
Funding
DS duly acknowledge the financial support from CSIR, Govt. of India for CSIR-Fast Track Translation (FTT) project (33/BS/FTT/2016-PPD) and CSIR Network Project CeHAB (BSC0209).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, V., Kumar, S., Darnal, S. et al. Optimized chromogenic dyes-based identification and quantitative evaluation of bacterial l-asparaginase with low/no glutaminase activity bioprospected from pristine niches in Indian trans-Himalaya. 3 Biotech 9, 275 (2019). https://doi.org/10.1007/s13205-019-1810-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1810-9