Abstract
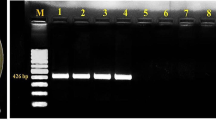
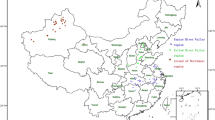
Thirty-four Xanthomonas citri pv. malvacearum (Xcm) isolates collected from three cotton-growing zones of India were subjected for virulence and race documentation and further correlated with genetic diversity as revealed by repetitive elements [repetitive extragenic palindromic (REP), enterobacterial repetitive intergenic consensus (ERIC) and BOX elements] and intersimple sequence repeat (ISSR)—PCR analyses. Among the 34 isolates tested for virulence on susceptible cultivar LRA 5166, 7 were recorded as highly virulent (HV), 16 were moderately virulent (MV) and 11 were less virulent (LV). Eight different races were recorded by using ten cotton host differentials. Twenty-two isolates (65%) belonged to race 18. Twelve isolates (35%) pertained to races 3, 5, 6, 7, 8, 11 and 13. REP, ERIC, BOX, combined repetitive elements, and ISSR analyses revealed the presence of 7, 10, 9, 11, and 8 clusters, respectively, at similarity coefficient of 0.70 in dendrograms. Principal coordinate analysis (PCoA) exhibited 76.4% and 77.5% cumulative variability for combined repetitive elements and ISSR analyses. ERIC produced the highest polymorphic information content (PIC) value (0.928). A lot of intra-pathovar variability was observed in virulence and genomic fingerprinting among Xcm isolates. Many of the isolates grouped based on geographical origin irrespective of virulence or race. The spread of the pathogen races in India might be due to the transport of germplasm lines and seed materials from one place to others.




Similar content being viewed by others
References
Abdo-Hasan M, Khalil H, Debis B, Ali-Mir N (2008) Molecular characterization of Syrian races of Xanthomonas axonopodis pv. malvacearum. J Plant Pathol 90:431–439
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Archana S, Prabukarthikeyan R, Raguchander T, Prabakar K (2017) Comparative assessment of RAPD and ISSR markers to study genetic polymorphism in Colletotrichum gloeosporioides isolates of mango. Asian J Plant Pathol 11:130–138
Asgarani E, Ghashghaei T, Soudi MR, Alimadadi N (2015) Enterobacterial repetitive intergenic consensus (ERIC) PCR based genetic diversity of Xanthomonas spp. and its relation to xanthan production. Iran J Microbiol 7:38–44
Chen W, Kuo T (1993) A simple and rapid method for the preparation of gram negative bacterial genomic DNA. Nucleic Acids Res 21:2260
Dangi AK, Sharma B, Khangwal I, Shukla P (2018) Combinatorial interactions of biotic and abiotic stresses in plants and their molecular mechanisms: systems biology approach. Mol Biotechnol. https://doi.org/10.1007/s12033-018-0100-9
Dombek PE, Johnson LK, Zimmerley ST, Sadowsky MJ (2000) Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl Environ Microbiol 66:2572–2577
El-Zik KM, Thaxton PM, Creech JB (2001) A pedigree analysis of cotton cultivars affected by bronze wilt. In: Proceedings of beltwide cotton conference, Anaheim, CA 9–13 Jan, 2001, Natl Cotton Counc Am, Memphis, TN, USA, pp 105–107
Follin JC, Girardot B, Mangano N, Benitez R (1988) New results on inheritance of immunity to bacterial blight, Xanthomonas campestris pv. malvacearum (Smith) Dye race 18 and 20 in cotton plant (Gossypium hirsutum L.). Coton et Fibres Tropicales 43:167–174
Gholve VM, Kurundkar BP, Meshram MK, Wasule DL (2005) Prevalence of races of Xanthomonas axonopodis pv. malvacearum in Marathwada region of Maharashtra State. J Cotton Res Dev 19:254–256
Hillocks RJ (1992) Bacterial blight. In: Hillocks RJ (ed) Cotton diseases. CAB Int, Wallingford, pp 39–85
Hunter RE, Brinkerhoff LA, Bird LS (1968) The development of a set of upland cotton lines for differentiating races of Xanthomonas malvacearum. Phytopathology 58:830–832
Imam J, Shukla P, Mandal NP, Variar M (2017) Microbial interactions in plants: perspectives and applications of proteomics. Curr Protein Pept Sci 18:1–10
Jalloul A, Sayegh M, Champion A, Nicole M (2015) Bacterial blight of cotton. Phytopathol Mediterr 54:3–20
Jensen BD, Vicente JG, Manandhar HK, Roberts SJ (2010) Occurrence and diversity of Xanthomonas campestris pv. campestris in vegetable Brassica fields in Nepal. Plant Dis 94:298–305
Klanderud K, Meineri E, Töpper J, Michel P, Vandvik V (2017) Biotic interaction effects on seedling recruitment along bioclimatic gradients: testing the stress-gradient hypothesis. J Veg Sci 28:347–356
Lopez R, Asensio C, Gilbertson RL (2006) Phenotypic and genetic diversity in strains of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) in a secondary center of diversity of the common bean host suggests multiple introduction events. Phytopathology 96:1204–1213
Louws FJ, Fulbright DW, Stephens CT, de Bruijn FJ (1994) Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol 60:2286–2295
Louws FJ, Rademaker J, de Bruijn FJ (1999) The three Ds of PCR-based genomic analysis of phytobacteria: diversity, detection, and disease diagnosis. Ann Rev Phytopathol 37:81–125
Madani AS, Marefat A, Behboudi K, Ghasemi A (2010) Phenotypic and genetic characteristics of Xanthomonas citri subsp. malvacearum, causal agent of cotton blight, and identification of races in Iran. Australas Plant Pathol 39:440–445
Massomo SMS, Nielsen H, Mabagala RB, Mansfeld-Giese K, Hockenhull J, Mortensen CN (2003) Identification and characterisation of Xanthomonas campestris pv. campestris strains from Tanzania by pathogenicity tests, Biolog, rep-PCR and fatty acid methyl ester analysis. Eur J Plant Pathol 109:775–789
Meshram MK, Sheo R, Wasule DL (2002) Bacterial Blight. In: Sheo R (ed) Twenty five years of achievements in cotton pathology at CICR (1976–2001). Central Institute for Cotton Research, Nagpur
Mhedbi-Hajri N, Hajri A, Boureau T, Darrasse A, Durand K, Brin C, Fischer-Le Saux M, Manceau C, Poussier S, Pruvost O, Lemaire C, Jacques M-A (2013) Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis. PLoS One 8:e58474
Moretti C, Vinatzer BA, Onofri A, Valentini F, Buonaurio R (2017) Genetic and phenotypic diversity of Mediterranean populations of the olive knot pathogen, Pseudomonas savastanoi pv. savastanoi. Plant Pathol 66:595–605
Nunes MP, Mehta A, Aguiar PH, Cia E, Pizzinato MA, Chiavegato EJ, Mehta YR (2009) Analysis of genetic diversity among the isolates of Xanthomonas axonopolis pv. malvacearum of cotton. Summa Phytopathol 35:105–109
Oliveira JC, Albuquerque GMR, Xavier AS, Mariano RLR, Suassuna ND, Souza EB (2011) Characterization Xanthomonas citri subsp. malvacearum causing cotton angular leaf spot in Brazil. J Plant Pathol 93:707–712
Patil PV, Patel JR, Patel UG (2003) Assessment of avoidable yield losses caused by bacterial blight in G. cot. Hy10 Cotton and its parents. J Cotton Res Dev 17:45–47
Rademaker JLW, Hoste B, Louws FJ, Kersters K, Swings J, Vauterin L, Vauterin P, de Bruijn FJ (2000) Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol 50:665–677
Raghavendra VB, Siddalingaiah L, Prakash HSH (2009) Role of cultivars, physical, chemical and organic treatments in the management of bacterial blight of cotton. Arch Phytopathol Plant Prot 42:1101–1108
Rohlf FJ (1997) NTSYS-PC Version. 2.02i, numerical taxonomy and multivariate analysis system. Applied Biostatistics Inc., Exeter Software
Samanta JN, Mandal K (2014) Assessment of intrapathovar variability of Xanthomonas axonopodis pv. commiphorae through phenotypic and molecular markers. For Pathol 44:85–95
Schaad NW, Jones JB, Chun W (2001) Plant pathogenic bacteria, 3rd ed. The Bacteriology Committee of the American Phytopathological Society. APS Press, St. Paul
Schaad NW, Postnikova E, Lacy G, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK (2007) Xanthomonas alfalfae sp. nov., Xanthomonas citri sp. nov. and Xanthomonas fuscans sp. nov. In: List of new names and new combinations previously effectively, but not validly, published, validation list no. 115. Int J Syst Evol Microbiol 57:893–897
Sheoraj (1988) Grading for cotton disease. Central Institute for Cotton Research, Nagpur, pp 1–7 (Bulletin)
Singh D, Rathaur PS, Singh A, Raghuwanshi R (2015) Genetic diversity of Xanthomonas campestris pv. campestris isolated from Brassica crops using RAPD and Rep-PCR. Int J Agric Sci 85:792–796
Singh D, Rathaur PS, Vicente JG (2016) Characterization, genetic diversity and distribution of Xanthomonas campestris pv. campestris races causing black rot disease in cruciferous crops of India. Plant Pathol 65:1411–1418
Thaxton PM, El-Zik KM (2001) Bacterial blight. In: Kirkpatrick TL, Rothrock CS (eds) Compendium of cotton diseases, 2nd edn. APS Press, St. Paul, pp 34–35
Thaxton PM, Brooks TD, El-Zik KM (2001) Race identification and severity of bacterial blight from natural infestations across the Cotton Belt. In: Proceedings of beltwide cotton conference, Anaheim, CA, 9–13 Jan 2001, National Cotton Council of America, Memphis, TN, USA, pp137–138
Trindade LC, Lima MF, Ferreira MASV (2005) Molecular characterization of Brazilian strains of Xanthomonas campestris pv. viticola by rep-PCR fingerprinting. Fitopatol Bras 30:46–54
Valverde A, Hubert T, Stolov A, Dagar A, Kopelowitz J, Burdman S (2007) Assessment of genetic diversity of Xanthomonas campestris pv. campestris isolates from Israel by various DNA fingerprinting techniques. Plant Pathol 56:17–25
Vauterin L, Rademaker J, Swings J (2000) Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology 90:677–682
Verma JP, Singh RP (1975) Studies on the distribution of races of Xanthomonas malvacearum in India. Indian Phytopathol 28:459–463
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831
Versalovic J, Schneider M, de Bruijn FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5:25–40
Vinatzer BA, Monteil CL, Clarke CR (2014) Harnessing population genomics to understand how bacterial pathogens emerge, adapt to crop hosts, and disseminate. Ann Rev Phytopathol 52:19–43
Zhai JL, Luo YH, Zheng DJ, Huang X (2010) Evaluation of genetic diversity of highly virulent strains of Xanthomonas campestris pv. malvacearum by rep-PCR fingerprinting. J Phytopathol 158:764–768
Acknowledgements
We acknowledge the support given by the Director, ICAR-Central Institute for Cotton Research, Nagpur, Professor and Head, Department of Plant Pathology and Dean (SPGS), Tamil Nadu Agricultural University, Coimbatore. We also extend our sincere thanks to DST (FIST) and UGC (SAP) for providing laboratory facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, A.S., Aiyanathan, K.E.A., Nakkeeran, S. et al. Documentation of virulence and races of Xanthomonas citri pv. malvacearum in India and its correlation with genetic diversity revealed by repetitive elements (REP, ERIC, and BOX) and ISSR markers. 3 Biotech 8, 479 (2018). https://doi.org/10.1007/s13205-018-1503-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1503-9