Abstract
The freshwater sponge, Ephydatia muelleri, is an emerging model system for studying animal:microbe symbioses. Intracellular green microalgae are one of the more common symbionts that live in a facultative mutualism with E. muelleri. While these symbioses have long been known, the identity of the algal symbionts in E. muelleri cells has not been studied in detail. Here, we isolate and characterize endosymbiotic algae from E. muelleri collected from different geographic locations. We find that the algae can be transmitted through asexually produced gemmules and importantly that they can form symbioses with different, differentiated sponge cell types in the adult sponge. Our findings indicate that at least two algal lineages form endosymbioses with E. muelleri. One of the lineages includes species commonly found in samples from two locations in Canada and one in the United States (clade 1: closely related to Auxenochlorella pyrenoidosa). The other clade includes algae found in sponges from one site in Maine, USA, and Lewiniosphaera symbiontica, which is a strain isolated in 1956 from the freshwater sponge Spongilla. We compared microbiomes found in cultures of microalgae as well as the original sponge hosts, and found that very similar bacterial microbiomes associate with both clades (91 orders of Bacteria are shared among the samples we compared). The microbiomes found in the cultures resemble, with a high degree of overlap, the microbiome associated with the sponge host.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Sponges are among the earliest branching animal phyla (Telford et al. 2016) and form associations with a large variety of microbial partners (Thomas et al. 2016; Pita et al. 2018). The holobiont (i.e., host plus associated symbionts) performs key functions in the ecosystem, and holobiont partners generate reciprocal selective pressures that result in interesting evolutionary outcomes (e.g., Taylor et al. 2007; Webster & Taylor 2012). Photosynthetic symbionts account for an important part of the holobiont both in marine and freshwater sponges. These symbionts provide photosynthates, secondary metabolites, and possibly are involved in nitrogen fixation to the sponge host (Wilkinson and Fay 1979; Wilkinson 1983; Arillo et al. 1993). Photosynthetic symbionts include cyanobacteria, dinoflagellates, rhodophytes, chlorophytes and diatoms (Scott et al. 1984; Rützler 1985; Taylor et al. 2007). Cyanobacteria are a common and well-described group of photosynthetic endosymbionts that can reside in marine and freshwater sponges (e.g., Thacker and Freeman 2012; Webster and Taylor 2012; Gaikwad et al. 2016; Kulakova et al. 2014) where they can play important roles in host adaptation through expanded metabolic function via photoautotrophy (e.g., Zhang et al. 2015; Hudspith et al. 2022). A small number of marine sponges, like some in the Clionaidae, possess photosynthetic dinoflagellate endosymbionts (Gerakladium) that can enhance bioerosion and growth rates (Hill 1996; Weisz et al. 2010; Ramsby et al. 2017). Co-occurrences of sponge:rhodophyte associations have been described where the organisms grow attached to one another producing surface proliferations (Tronchin et al. 2006) and some of the most abundant diatom species of the Antarctic plankton communities have been observed in high densities inside the tissues of several sponge species (Cerrano et al. 2004). Last but not least, and the focus of this study, is the long-known relationship of chlorophytes, also known as green algae, that form endosymbioses with freshwater sponges (Brandt 1882).
Algal photobionts are abundant in freshwater sponges of the Demospongiae, but less is known about this intriguing partnership than of marine sponges. The symbiotic intracellular algae of freshwater sponges include a diverse set of chlorophytes from a variety of genera mostly within the class Trebouxiophyceae (Kenney et al. 2019; Ereskovsky et al. 2022). Many of the reports of endosymbiotic green algae have been from the freshwater sponge, Spongilla lacustris (e.g., Castro-Rodriguez 1930; Gilbert and Allen 1973; Williamson 1979; Reisser 1984; Saller 1989, 1991; Masuda 1990; Handa et al. 2006). Symbiotic algae in S. lacustris and in Radiospongilla cerebellata have been shown to affect germination rates of gemmules (Agnes and Brøndsted 1953; Okuda et al. 2002) and have a positive effect on the growth rate of sponges due to net gain from photosynthesis (Frost and Williamson 1980; Sand-Jensen and Pedersen 1994). Additionally, Cook (1983) demonstrated that algal symbionts benefit another freshwater sponge species, Ephydatia fluviatilis, by providing glucose to host sponge cells, and by providing up to 20% of the total fixed carbon for the host’s metabolism.
For the majority of freshwater sponge symbioses involving green algae, the precise identity of the partners is unknown. Chlorella sorokiniana is known to be a species of algae that is associated with Spongilla (Reisser 1984), but most studies have not identified the species of algae using molecular markers. For example, freshwater sponges with documented green algae of the Trebouxiophyceae include Radiospongilla sp. (Masuda 1990; Handa et al. 2006), Eunapius fragilis (Handa et al. 2006), Lubomirskia sp. (Berner and Titlyanov 1992; Bil et al. 1999; Ereskovsky et al. 2016), E. fluviatilis (Wilkinson 1980; Gaino et al. 1995), and E. muelleri (Hall et al. 2021). One study showed that gemmules from R. cerebellata possessed two different endosymbiotic algal species (Handa et al. 2006). In most of the cases described above, morphology was used for taxonomic classification. However, more recent genetic sequence data has become available for algae found in Lubomirskia and Baikalospongia freshwater sponge species; the symbionts in these sponges were identified as belonging to the coccoid green algal genus, Mychonastes in the case of Lubomirskia baicalensis (Chernogor et al. 2013) and multiple lineages of Choricystis in the case of several Lubomirskia and Baikalospongia sponge species in Lake Baikal (Kulakova et al. 2020). Pröschold and Darienko (2020) recently analyzed a symbiotic strain of S. lacustris isolated by Lewin (1966) in Massachusetts, USA and determined it to be a member of the Chlorellaceae representing a new species of the genus Lewiniosphaera (L. symbiontica) using both morphological and DNA sequence analysis. In spite of abundance and importance of freshwater sponge:algal symbioses, relatively little is known about the Chlorella-like algae that form endosymbiotic relationships with freshwater sponges.
Recently, Ereskovsky et al. (2022) reviewed the literature on cytosymbiosis and algal symbionts in sponges. They discuss the evidence that adult stem cells in sponges (most frequently archaeocytes) host endosymbionts and that this arrangement is contrasted with the observation that intracellular symbionts commonly reside in more specialized or differentiated cells in other animal hosts. They make the argument that Porifera should be a model for studying host:symbiont coevolution because sponge holobionts are ancient symbiotic associations among the Metazoa and because adult stem cell cytosymbioses are rare. We agree that sponges, particularly the freshwater sponges, offer unique opportunities to study animal:algal endosymbioses. We recently showed that the freshwater sponge, E. muelleri, is a tractable model for studying sponge:algal endosymbiosis (Hall et al. 2021). Ephydatia muelleri is nearly panglobal, is globally abundant, and easy to culture in the lab with and without symbionts. This species of sponge has a well-annotated chromosomal level genome and a developmentally staged transcriptome (Kenny et al. 2020). E. muelleri has also been an important species for studying physiology, cell biology, and genetics of sponges, especially in the context of animal evolution (Elliott and Leys 2007, 2010; Windsor & Leys 2010; Ludeman et al. 2014; Schenkelaars et al. 2016; Windsor-Reid et al. 2018; Hall et al. 2019; Mitchell and Nichols 2019; Colgren and Nichols 2022). Several transcriptomes representing various stages of symbiosis (Hall et al. 2021; Geraghty et al. 2021) have been analyzed, which revealed a set of sponge genes involved in establishment and maintenance of the symbiosis. However, algal-specific gene expression or the taxonomic identity of the native Chlorella-like algal symbiont in E. muelleri has not been analyzed. In an effort to better understand these important symbioses, we present morphological and molecular data describing E. muelleri algal and microbial endosymbionts isolated from several regions in Canada and the USA.
2 Materials and methods
2.1 Algal strain isolation and culturing
Ephydatia muelleri tissue possessing microalgal symbionts was collected from several different locations in the USA and Canada and algal symbionts were isolated from green E. muelleri freshwater adult sponges or gemmules (Table S1). As adult sponges can possess both endosymbionts and other associated algae, to ensure that only intracellular endosymbionts were isolated (not free-living algae), gemmules or hatched gemmules were used when available. In the case of algae that were isolated from adult sponges, to ensure strains were pure, algae were cultured on agar plates as follows. Algae were isolated by grinding tissue followed by differential centrifugation as described in Hill et al. (2020). Algal cultures, both xenic and axenic, were grown in both Bold 3N medium (UTEX Culture Collection of Algae) and Bold Modified Basal Freshwater Nutrient Solution (BBM, Sigma Aldrich). All liquid cultures were grown at 20–25 ℃ under a light:dark cycle of 12:12 h for as long as 6 weeks before periodic subculture. To remove unwanted microorganisms from the aqueous cultures and to ensure strains were pure, algae were streaked on BBM agar plates for several rounds until single strains were isolated. Frozen stocks (-80 ℃) of axenic strains were made using the GeneArt Cryopreservation Kit for Algae (Invitrogen).
2.2 Freshwater sponge culturing
Ephydatia muelleri gemmules were collected, stored, and cultured as described in Leys et al. (2019). Gemmules are overwintering, asexually produced small cysts of sponge stem cells that are capable of developing into a juvenile sponge upon hatching. Gemmules possessing high amounts of algae inside the gemmule covering and among the stem cells were green and could be used for algal isolation or for hatching sponges with algal endosymbionts in further experiments. Alternatively, aposymbiotic sponges hatched from gemmules could be infected with algae to establish the symbiosis according to Hill et al. (2020). Sponges and associated algal symbionts were either harvested and stored at -20o C for later DNA isolation or grown on 35 mm glass bottom culture dishes (MatTek Life Sciences) and fixed in cold 4% paraformaldehyde (PFA) in 190 proof ethanol overnight for microscopy.
2.3 Microscopy
Algae and associated microorganisms grown on agar plates were imaged using a Leica M165C stereoscope with Leica MC170 HD camera. Algae in liquid cultures were concentrated by centrifugation at 3000 rpm for 10 min to obtain a pellet that could be resuspended in algal media and mounted in 100% glycerol for morphological analysis using brightfield microscopy with a Nikon 80i fluorescence upright microscope at 100 × oil immersion objective. Using the same microscope and lens, images of sponge cells and associated microalgae from ruptured gemmules were obtained by placing a single gemmule on a microscope slide and either squashing with a coverslip or tearing with forceps and then placing a small amount of glycerol on the expelled cellular contents followed by a coverslip.
Sponges with symbionts that were fixed in 4% paraformaldehyde (PFA)/ethanol were washed three times in phosphate buffered saline (PBS), treated with phosphate buffered saline with 1% Tween 20 (PBST) for 45 min, washed again in PBS, and stained with Alexa Fluor™ 488 Phalloidin (1:40 dilution, Invitrogen, Molecular Probes) and Hoechst 33342 (1:2000 dilution, Thermo Scientific). Samples were imaged using the Leica SP8 laser scanning confocal microscope equipped with white light laser (WLL) system, hybrid detectors and 63 × water immersion objective.
2.4 DNA extraction, molecular barcoding, and phylogenetic analysis
Genomic DNA of algal cultures was extracted using CTAB reagent and mechanical disruption with 0.5 mm Zirconia/Silica beads and the Bead Ruptor 4 (Omni International). The SSU rRNA and chlorophyte genes were amplified using GoTaq PCR 2X MasterMix (Promega). Chlorophyte gene marker amplifications were subject to the following thermocycling conditions: initial denaturation at 95 °C for 5 min, followed by 34 cycles of 95 °C for 30 s, 53 °C for 30 s, 72 °C for 1 min 20 s, followed by 1 cycle of 72 °C for 5 min. SSU rRNA gene marker amplifications were subject to the following thermocycling conditions: initial denaturation at 95 °C for 3 min, followed by 34 cycles of 95 °C for 1 min, 55 °C for 1 min, 72 °C for 3 min, followed by 1 cycle of 72 °C for 5 min. SSU rRNA primers used included EAF3, E528F, 920F, GF, GR, and ITS055R (Marin et al. 1998, 2003) and newly designed primers as shown in Table S2. Chlorophyte gene markers used included TUFA, ITS, 16S and RBCL (Burja et al. 2001; Fama et al. 2002; Sun et al. 2009; Bock et al. 2010; Zou et al. 2016, Table S3). All PCR products were purified using QIAquick MinElute PCR Purification Kit (Qiagen) and sequenced. All sequences are provided in File S1.
As morphology was not informative for the taxonomic identification of the algal isolates, we implemented a molecular taxonomy approach to decipher whether individuals of E. muelleri from different geographical locations possessed the same or different endosymbiotic Chlorella-like algae. For each geographic isolate shown in Fig. 1 and for an isolate from Virginia that was originally described in Hall et al. (2021), we sequenced three commonly used molecular markers for algal taxonomy: two chloroplast markers—the elongation factor Tu gene (tufA) and the large subunit of the enzyme ribulose bisphosphate carboxylase (RuBisCo) (rbcL)—and the nuclear marker ribosomal small subunit DNA (SSU rDNA) including the internal transcribed spacer (ITS) between the small and large-subunit ribosomal (rRNA).
Morphology of Symbiotic Algal Isolates of Ephydatia muelleri. A. O’ Connor Lake, British Columbia, Canada (Emu_LOC). Isolated from gemmules. B. Saint Lawrence River, Montreal, Canada (Emu_MON). Isolated from adult tissue. C. Pemaquid River, Maine, U.S. (Emu_PR6). Isolated from adult tissue. D. Sooke Reservoir, British Columbia, Canada (Emu_Sooke). Isolated from adult tissue. E. Dundee Pond, Maine, U.S. (Emu_DPAG). Isolated from hatched gemmules. F. Pemaquid River, Maine, U.S. (Emu_PRAG). Isolated from gemmules and hatched gemmules. Scale bar for all images = 10 µm
Known algal sequences were obtained from the NCBI database nucleotide collection (nr/nt) and aligned with our SSU rRNA and chlorophyte sequenced genes separately using MAFFT (Katoh and Standley 2013). Phylogenetic trees were built with the chloroplast genes tufA and rbcl and the nuclear SSU-ITS concatenated using maximum likelihood in IQ-TREE (Nguyen et al. 2015; Minh et al. 2020) under the TIM2e + R3 model. All models were tested by ModelFinder (Kalyaanamoorthy et al. 2017) and the optimal was selected based on Bayesian Information Criterion (BIC) and Akaike Information Criterion (AIC) statistics Branch support was assessed using the ultrafast bootstrap (UF bootstrap) method (Minh et al. 2013).
2.5 Algal microbiome sequencing and analysis
Total genomic DNA of each of the xenic algal isolates was also used to complete a 16S rRNA gene microbiome analysis. A 464 bp hypervariable region of V3 (ONT_unipro341F) and V4 (ONT_unipro805R) of bacterial and archaeal DNA (Takahashi et al. 2014; Apprill et al. 2015; Parada et al. 2016) was amplified using LongAmp Taq 2X Master Mix (New England BioLabs). Thermocycling conditions for the first round of PCR included initial denaturation at 98 °C for 2 min, followed by 10 cycles of 95 °C for 20 s, 65 °C for 15 s, -1 °C/cycle, 65 °C for 30 s, followed by 25 cycles of 95 °C for 20 s, 55 °C for 15 s, and 65 °C for 1 min and a final extension at 1 cycle of 65 °C for 5 min. The PCR Barcoding Expansion Pack 1–96 (EXP-PBC096, Oxford Nanoporetech, US) was used to barcode PCR libraries with thermocycling conditions of 16S rRNA gene amplicons with an initial denaturation at 95 °C for 3 min, followed by 15 cycles of 95 °C for 15 s, 62 °C for 15 s, 65 °C for 1 min, followed by 1 cycle of 65 °C for 5 min. The Ligation Sequencing Kit 1D (SQK-LSK109, Nanopore Technologies) and the NEBNext® Companion Module for Oxford Nanopore Technologies® Ligation Sequencing (E7180S, New England BioLabs) were used following the manufacturers’ protocols. The barcoded gene libraries were pooled in equal amounts of DNA and sequenced in a MinION Flow Cell for ~ 5 h. The MinKNOW report containing the aggregated counts tables at different taxonomic levels was generated by the cloud-based Epi2ME software (Oxford Nanopore, USA). Following analyses were performed in R (version 4.2.1).
2.6 Sponge microbiome sequencing and analysis
DNA was extracted from about 20 mg of tissue using the DNeasy Blood and Tissue kit (Qiagen). The V4 hypervariable region was amplified with the same primers used for the algae samples, 515F-Y (Parada et al. 2016) and 806R (Apprill et al. 2015). DNA amplification was done in duplicates with the following conditions: 95 °C for 20 s, followed by 25 cycles of 95 °C for 10 s, 60 °C for 20 min, 72 °C for 30 min, and a final elongation at 72 °C for 5 min. Libraries were prepared with the Nextera XT DNA Library Preparation Kit (Illumina Inc.) and next generation paired-end sequencing was performed at the Natural History Museum of London (https://www.nhm.ac.uk/) on an Illumina MiSeq device using v3 chemistry (2 × 300 bp). Read processing and taxonomic assignment followed the MiSeq SOP protocol (Kozich et al. 2013) in Mothur (v.1.41.3) inferring amplicon sequence variants (ASVs), allowing one mismatch per 100 bp. ASVs were classified using the Reference NCBI database (refseq_rna, update 12/01/2023, including 49,700,764 sequences), with a cutoff value of 80.
2.7 Statistical analyses of algal and sponge microbiomes
Measures of alpha diversity (Shannon index) were calculated using rarefied samples in R. These metrics were compared among clades or locations using analyses of variance (ANOVA), and Tukey's honest significant difference (HSD) for pairwise comparison. Beta-diversity among the microbiome samples was visualized using principal coordinate analysis (PCoA) on a Bray–Curtis dissimilarity matrix using "cmdscale" in vegan package v. 2.5-7 (Oksanen et al. 2018). Homogeneity of variance, which tests whether two or more groups are homogeneously dispersed in relation to their group centroid, was determined using the "betadisper" function in vegan. We compared distances among clades and locations by permutational multivariate analyses of variance (PERMANOVA) using “adonis” in vegan.
3 Results
3.1 Morphology and cellular location of E. muelleri algal symbionts
Both endosymbiotic and putatively ectosymbiotic algal strains were identified in E. muelleri tissues and gemmules. In all cases, endosymbiotic algae were culturable in modified Bold’s media and, regardless of geographic location, the predominant algae were unicellular zoochlorellae, small (4–10 µm), spherical, and without flagella (Fig. 1). As has been described in other work on green algal symbionts in freshwater sponges, these algae appear to be Chlorella-like in this facultative symbiosis. We did not observe significant morphological differences between the strains.
We considered whether algal symbionts transmitted to sponges through gemmules were present inside or outside of host thesocytes (sponge stem cells in the gemmules). We examined the cellular contents of E. muelleri gemmules to evaluate the location of algae prior to thesocyte differentiation (i.e., thesocytes are the resting state of archeocyte stem cells in the gemmule) and sponge development to determine where algal cells resided (Fig. 2). We observed that even ‘yellow’ gemmules (those without obvious symbionts) possessed some algal cells, mostly located outside of thesocytes (Fig. 2A). ‘Green’ E. muelleri gemmules possessed greater numbers of algae and while most of the algal cells were located outside of sponge cells, there were thesocytes that appeared to contain intracellular algae (Fig. 2B and C). Thus, it is possible that the algae persist in those thesocytes and are passed onto future cells through cell division. Gemmules were observed to contain diverse communities of bacteria.
Ruptured gemmules cellular content. A. Cells from a ruptured yellow E. muelleri gemmule with inset of yellow gemmules with two popped to show cellular contents. B. Cells from a ruptured green E. muelleri gemmule with inset of green gemmules. C. Stem cell (thesocyte) with putative intracellular algae shown by arrowhead. Scale bar for gemmule cells = 10 µm
We hatched E. muelleri gemmules to examine algal symbiont location in sponges developing from gemmules. Using confocal microscopy, we verified that all strains of algae isolated from E. muelleri gemmules or hatched gemmules (Table S1) were present in intracellular locations in juvenile sponges. E. muelleri hatched from ‘green’ gemmules collected from three of the geographical locations contained algae in intracellular spaces (Fig. 3A-C) in multiple cell types. Depending on the number of algae in the gemmule and the length of time the sponge is grown with the algae, the numbers of algae per sponge cell varied, with dark green gemmules grown to adult sponges having numerous algae per cell (Fig. 3A and B). Sponges grown from gemmules possessing few algae (‘yellow’ gemmules) had fewer endosymbionts (Fig. 3C), but our earlier work showed that when sponges are grown under light, numbers of algae increase over time as the sponge and algal cells divide (Hall et al. 2021).
E. muelleri cells with endosymbiotic algae. A. Panel A-C shows sponge tissue grown from green gemmules collected from different geographical locations. A. O’ Connor Lake, British Columbia, Canada. B. Dundee Pond, Maine, U.S. C. Pemaquid River, Maine, U.S. Panel D shows E. muelleri tissue grown from “aposymbiotic” gemmules and infected with the algal endosymbiont strain isolated from gemmules obtained from that geographic location. D. Pemaquid River, Maine, U.S. infected with native algal strain, 20 min post infection. Tissue stained with Phalloidin 488 for actin (green) and Hoescht for nuclei (white in panel A-C., blue in panel D.) Autofluorescence of algae is observed (red). Scale bar for all images = 25 µm
We have previously demonstrated (Hall et al. 2021) that E. muelleri hatched from aposymbiotic gemmules (or gemmules with very low numbers of algae) can be infected with symbiotic algal strains isolated from the same geographic location and that these endosymbioses can be maintained in the lab for as long as the sponges are viable. Here, we verified this finding using strains of algae and E. muelleri from new geographical locations. Figure 3D shows aposymbiotic E. muelleri hatched from gemmules from the Pemaquid River (Maine) infected with the Emu_PR6 native algal strain. While some algae remain in extracellular locations, 20 min after infection the majority of algae are intracellular.
3.2 Molecular barcoding and phylogenetic analysis of algal symbionts
To place our algal sequences within known algal clades, we collected the most similar sequences to those produced herein from the best hits resulting from BLAST analysis against the nr database from NCBI (File S1). All three strains from Canada (LOC, MON, SOOKE), one strain from Maine (DPAG), and the one from Virginia (VA SI) resulted in similar outputs. Using the nuclear marker SSU, the most similar sequences were those from Pseudochlorella pringsheimii. The chloroplast marker gene tufA for our strains blasted to Auxenochlorella pyrenoidosa. The chloroplast marker rbcL was not particularly informative for the classification of these queries as it resulted in multiple best hits (Auxenochlorella pyrenoidosa, Jaagichlorella luteoviridis, Chlorella vulgaris, Auxenochlorella protothecoides, and Chlorella pyrenoidosa). The other two algal strains studied here, PR6 and PRAG, both isolated from individuals of E. muelleri from Permaquid River (Maine), blasted against different algal strains than LOC, MON, SOOKE, DPAG and VA SI. The SSU queries for PR6 and PRAG were notably identical to the newly erected species of Lewiniosphaera symbiontica that was recently described as an isolate from a freshwater sponge endosymbiont of Spongilla collected in Massachusetts in 1956 (Pröschold and Darienko 2020). The tufA marker revealed the closest similarity for these strains with Chlorella sp. and the rbcL marker with Chlorella variabilis.
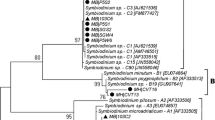
For a more accurate taxonomic assignment and in order to determine possible evolutionary relationships between the algal isolates from this study and other described algal species, maximum likelihood phylogenetic trees were built for all the molecular markers. The nuclear markers SSU and ITS were concatenated and a phylogenetic tree was reconstructed based on the SSU-ITS tree reported in Pröschold and Darienko (2020). The alignment for SSU-ITS included 46 algal sequences with 2292 nucleotide positions, 456 distinct patterns, 334 parsimony-informative sequences, 220 singleton sites, and 1738 constant sites. The algal isolates from Virginia (VA SI), Canada (LOC, MON, SOOKE) and the strain DPAG from Maine (DPAG) clustered together in a clade with Micractinium tetrahymenae with the highest bootstrap support, while the other two strains from Maine (PR6 and PRAG) formed a highly supported clade with L. symbiontica (Fig. 4).
Molecular phylogeny of Chlorellaceae based on the concatenated aligned sequences of the nuclear markers SSU and ITS. The tree is inferred by maximum likelihood and the nodal supports are ultrafast bootstraps. The reference sequences are based on the dataset from Pröschold and Darienko (2020). The algal isolates from this study are in bold. Note that only bootstrap values over 70 are shown
For the tufA phylogeny (Fig. S1), a dataset of 39 complete tufA sequences from different Chlorella-like algal species was compiled, with 741 nucleotide positions, 319 distinct patterns, 292 parsimony-informative sequences, 35 singleton sites, and 414 constant sites. Here, our sequences for the E. muelleri algal strains were clustered together in similar clades as those recovered in the SSU-ITS phylogeny (Figs. 4, S1). While the strains VA SI, LOC, MON, SOOKE, and DPAG were placed in a clade with A. pyrenoidosa (clade 1), the strains PR6 and PRAG branched in clade 2 with two sequences of the Chlorella sp. strain 484 (Fig. S1).
Finally, to reconstruct the rbcL phylogeny (Fig. S2), we compiled a final dataset of 46 complete and partial sequences from different species of Chlorellaceae, with 1293 positions, 589 distinct patterns, 427 parsimony-informative, 94 singleton sites, and 772 constant sites. In the resulting maximum likelihood hypothesis, we observed the algal strains organized in two distinct phylogenetic clades (1 and 2) as with the previous markers with identical affinities. The strains PR6 and PRAG clustered here in a robustly supported clade (clade 2) with sequences from Chlorella sp. and Chlorella variabilis strains (Fig. S2). The strains VA SI, LOC, MON, SOOKE, and DPAG were recovered as a single clade (clade 1) with algal species from several genera including Auxenochlorella, Jaagichlorella and Pseudochlorella (Fig. S2).
3.3 Algal strain associated microbiomes
We found that the algae grew well, some strains for nearly a decade, in media or on plates with the microbes that were co-isolated from the sponges. Each of the strains of symbiotic algae isolated from either adult freshwater sponge tissue or from gemmules or hatched gemmules contained associated prokaryotic microorganisms (Fig. 5). We also found that these strains can be made axenic by colony isolation and multiple rounds of successive plating. The axenic strains also grow well in lab cultures, although most grow approximately 1.5 times slower than the xenic strains.
To better understand the diversity of bacteria we observed in algal cultures, we sequenced the microbiomes from endosymbiotic microalgal strains that were isolated from E. muelleri sponges and grown in culture for months to years with culture transfer approximately every 50 days (Table S1). Through long-read 16S rRNA gene amplicon sequencing we identified a total of 2,616 prokaryotic species (666 species with > 0.01 relative abundance). The identified species spanned 850 genera, 139 orders, 57 classes, and 26 phyla. We examined the bacterial community composition of the algal associated microbiomes using relative abundance plots at different taxonomic levels. At the phylum level, Cyanobacteria was the most abundant phylum in all samples followed by Proteobacteria (Fig. 6). These two phyla accounted for about 82% of the observations. Among Cyanobacteria, species from the orders Nostocales, Synechococcales, Oscillatoriales, and Pleurocapsales were present in all samples (Fig. 6). There were also four abundant Alphaproteobacteria orders and two Gammaproteobacteria orders shared among all samples (Fig. 6). Relatively high abundance of species from the orders Burkholderiales, Flavobacteriales, Mycoplasmatales, and Eubacteriales was observed in some but not all samples (Fig. 6). The microbiomes of the algal cultures isolated from sponges collected in Maine (PR6, PRAG and DPAG) were more similar to each other than to the rest of the microbiomes, regardless of the algal strains belonging to both clades (Fig. 6).
After identifying the most abundant taxonomic groups in the algal associated microbiomes, we asked whether there were different diversity patterns among the samples. The alpha diversity (ShannonH) ranged from 1.7 to 2.9, but it was not significantly different between clades (p-value = 0.34) or location (p-value = 0.44) likely due to a large variability among the samples (Fig. 7A and B). Considering only CA (SOOKE and LOC) and ME (PRAG, DPAG, and PR6), there was marginally significant differences (p-value = 0.06).
Diversity and PCA Analysis. A-B. Shannon diversity for samples arranged by clade (A) and location (B). C-D. Principal Coordinate Analysis (PCoA) plots of beta diversity of the algal microbiomes based on Bray–Curtis dissimilarity at species level. Only the host algae geographic location was a statistically significant grouping factor. Note that samples SOOKE and LOC are grouped in CA-VI, and PRAG, DPAG, and PR6 in ME
To understand the structure in the composition of the microbial communities, we ordinated the samples using a principal coordinate analysis (PCoA) based on Bray Curtis dissimilarity (Fig. 7). The first two coordinate axes explain 73.6% of the total variance observed in our sampling (Fig. 7C and D). The ordination plot revealed that the samples have a tendency to group by geographic location, however, the phylogenetic relationship of the host (algal clades) does not have a strong influence in the microbial assemblage. PERMANOVA test showed significant clustering by location (p = 0.005), but not by sponge clade (p = 0.14). Assumption of homogeneity of dispersion within groups was not refuted (p-value > 0.1).
These algal culture samples were compared to the microbiome of the adult sponges originally collected from three of the same locations as the algal isolates: CA-VI (which includes Sooke River (SOOKE) and Lake O’Connor (LOC)) and Maine (ME, Pemaquid River or PR). The sponge sample PR6_Em was the host from which the algal strain PR6 was sequenced. Sponge samples were sequenced with a different technology (File S1), therefore only a gross comparison at taxonomic levels of phylum and order was made. Similar to the algal samples, Cyanobacteria was the dominant group in the sponge microbiome (except for 3 samples in SOOKE), followed by Proteobacteria (Fig. 8). Many sponge samples, however, were characterized by high abundance of Bacteroidetes, that were not especially abundant in the algal microbiomes (Fig. 8). Similarly, sponge samples harbored medium abundances of Actinobacteria, Verrucomicrobia and Spirochaetes, that were decreased in the algal samples (Fig. 8A). Interestingly, similar to what occurs with the algal culture microbiomes (Figs. 6 and 8), sponge microbiomes from SOOKE were more similar to those collected in O’Connor Lake. At order level, the microbiomes of sponges and algae shared 91 bacterial orders (Fig. 8B), that included Microccocales (Actinobacteria), Chitinophagales and Flavobacteriales (Bacteroidetes), Pseudonabaenales, Nostocales, and Synechococcales (Cyanobacteria), Hyphomicrobiales, Rhodospirillales, Caulobacterales, Rhodobacterales, and Sphingomonadales (Alphaproteobacteria), and also Eubacteriales (Firmicutes).
Comparison of microbiota from algal isolates and sponges. A. Community composition comparison of the microbiota in the algal isolates and the sponge microbiome from E. muelleri at phylum taxonomic level represented as heatmap of log (relative abundance) and dendrogram calculated over dissimilarities at species level. B. Venn diagram showing the number of bacterial and archaeal orders shared by the samples of algal cultures and sponge hosts compared
4 Discussion
Endosymbionts found in E. muelleri from multiple locations in North America are green, coccoid, Chlorella-like microalgae (Fig. 1) that reside in a variety of sponge cells (Fig. 3). The transmission of algal symbionts in freshwater sponges likely occurs both horizontally, through ingestion of algae from the environment, and vertically during fragmentation, budding, or gemmulation (recently reviewed in Ereskovsky et al. 2022). To our knowledge, algae have not been observed in eggs or larvae of freshwater sponges. Whether or not algal symbiont transmission from asexual gemmule to adult sponge occurs via arrested phagocytosis of algal cells in differentiating thesocytes (i.e., stem cells stored in overwintering gemmules) or from algal symbionts that are already endosymbiotic in thesocytes is unknown. We suspect that both could happen given the fact that some of the algal strains we isolated were from gemmules or hatched gemmules that had been washed in hydrogen peroxide to kill microbes on the gemmule surface, and we found algae in the gemmule cyst both inside and outside of thesocytes (Fig. 2). While some algae in gemmules may be phagocytosed resulting in aposymbiotic sponges or sponges that do not show signs of algal division (e.g., Rasmont 1970; Simpson 1984), other algae avoid phagocytosis and persist in sponge cells (Fig. 3, Hall et al. 2021).
4.1 Algal identities
There is a moderate diversity of genera associated to freshwater sponges, which include Choricystis, Lewiniosphaera, and Chlorella, all notoriously difficult to identify based on their morphology. In this sense, several authors have used molecular markers to identify endosymbiotic algal species coming from sponge tissues in the last decades (e.g., Annenkova et al. 2011; Pröschold et al. 2011; Kulakova et al. 2014, 2020; Pröschold and Darienko 2020). In E. muelleri, we identified at least two species of green coccoid algae that can form an endosymbiosis with host sponge cells. We found that both species belong to the Chlorellaceae with the predominant species found in Canada and the U.S. to be closely related to Micractinium, Chlorella, Auxenochlorella, and Pseudochlorella genera, with the definitive identity varying depending on the molecular marker and available data set used. The SSU-ITS phylogenies revealed that the closest relative to our isolated algal strains from Canada, Virginia and from Dundee Pond in Maine (clade 1 in all trees) is Auxenochlorella pyrenoidosa, a previously known algal endosymbiont. Although the genus Auxenochlorella has not been described to live within sponges before, a strain of Auxenochlorella was identified from Hydra viridis tissue (Darienko and Pröschold 2015). Based also on SSU-ITS data, we suggest that the algal strains belonging to clade 2, which were isolated from E. muelleri gemmules and an adult sponge from the Pemaquid River in Maine, are Lewiniosphaera symbiontica, a species described by Pröschold and Darienko (2020) that was originally isolated in 1956 in Massachusetts in endosymbiosis with Spongilla lacustris. It is interesting to note that both PRAG and L. symbiontica have a 465 bp deletion in the 5’ region of the SSU-ITS. While we do not have additional sequences from L. symbiontica for other genes we sequenced (TufA, rbcL, 16S), the sequences for PRAG and PR6 are identical for those markers (Figs. 4, S1, and S2).
We also isolated strains of green algae from E. muelleri adult tissue (and not gemmules) that are not likely endosymbionts, but algae associated with the sponge tissue or from the water surrounding and within the sponge. We did not find these algae in intracellular locations, but consistently found these strains associated with E. muelleri at two geographical locations in Maine (Dundee Pond and Pemaquid River). The SSU/ITS marker showed that each of these strains belong to the Scenedesmaceae and are most closely related to Scenedesmus, Desmodesmus, and Tetradesmus algal species (File S2). We do not know if these algae play roles in the sponge trophic biology, but they are consistently present in samples of E. muelleri sponges we collect from these locations.
Given that E. muelleri can live with and without their algal endosymbionts and that the algae can live inside the host cells or outside of the animal, we believe that this facultative symbiosis provides a good model for further study regarding the mechanisms that regulate the symbiotic relationship, the evolution of host:symbiont specificity, as well as the geographical factors that might influence sponge-microbe mutualisms.
4.2 Algal microbes
While it is well known that sponges have complex microbiomes, so far, the nature of the microbes that may be associated with their algal endosymbionts has not been explored. Algae and bacteria can influence each other's metabolism, physiology, and growth through nutrient exchange, and they can mutually influence aquatic ecosystems including relationships between symbiotic algae and their animal hosts (reviewed in Ramanan et al. 2016).
Microalgae are surrounded by a phycosphere, a region rich in organic material made by the alga that serves as an association/exchange network for other organisms, like bacteria, to interact (Cirri and Pohnert 2019). Microalgal-bacteria interactions that influence algal growth and biomass production are supported by an array of compounds produced by bacteria living in communities together with algae (Fuentes et al. 2016) and it has been suggested that the positive effects of algal–bacterial interactions on algal growth should influence future research to move beyond consideration of algal associated bacteria as only contaminants (Astafyeva et al. 2022). We observed that algal endosymbionts of E. muelleri grow well in cultures that include bacteria that are co-isolated along with the algae from either adult sponge cells or from gemmules or hatched gemmules. We also found that it was difficult to isolate some of the algae from their associated bacteria growing in cultures (Fig. 5) coming from poriferan sources (i.e., multiple rounds of colony isolation on agar plates were needed to get axenic cultures). In fact, we frequently observed bacterial/yeast colonies growing in predictive patterns with the algae (Fig. 5) and documented algae growing on top of the other microbes in culture.
4.3 Algal culture and sponge associated bacteria
Recent studies in Hydra indicate that interactions of the Chlorella algal symbiont and extracellularly located microbiota shape the host microbiome (Bathia et al. 2022). The possibility that associated bacteria are important to the algal symbionts or to the endosymbiosis within sponges led us to characterize the algal microbiomes of each endosymbiont strain isolated from E. muelleri collected from different geographical locations (Fig. 6). To our knowledge, our study is the first to report the microbiota of endosymbiotic Chlorella-like algae isolated directly from environmental samples. A recent study has characterized the prokaryotic microbial community structure in Chlorella vulgaris SAG 211–12 cultures under different cultivation conditions in biotechnological systems (Haberkorn et al. 2020). In Haberkorn et al. (2020), the described bacterial community has originated from non-sterile handling of the pre-culture thus represents contamination that does not affect the algal growth in biotechnological reactors under different cultivation conditions. On the contrary, the algae prokaryotic community described here is representative of the microbial community that endosymbiotic Chlorella-like algae harbor in the wild.
In our study, each algal isolate possessed a unique microbiome signature, although there were some microbial families that were common to all microbiomes studied, and even to the microbiome of the host sponges (Figs. 6 and 8). The main phyla associated with the microbial communities present in endosymbiotic algal cultures isolated from E. muelleri were Cyanobacteria and Proteobacteria; though Bacteroidetes, Actinobacteria, Firmicutes, Tenericutes, and Euryarchaeota among a few others were also predominantly observed, but at lower frequencies. Some of the main families found in these microbiomes (i.e., Comamonadaceae, Caulobacteraceae, Chitinophagaceae, and Sphingomonadaceae) were also revealed from 16S sequencing of cultured strains of Chlorella saccharophila (Krohn-Molt et al. 2017). We acknowledge that long term cultivation of some of our strains (i.e., VA-SI, MON) may have resulted in inadvertent selection of distinct bacterial communities, but we believe that this baseline microbiome data provides information about microbes that can be associated with these endosymbiotic strains. The diversity patterns of the microbiomes of the algal cultures where independent of the identity of the algae, and they seem to be closely associated to either the geographical site and/or the host from which the algae where isolated (Fig. 7 and 8). This could indicate a strong influence of the environmental prokaryotes from which the host select their microbiomes, since these are Low Microbial Abundance sponges, with their microbiomes highly influenced by their horizontal acquisition strategy (Díez-Vives et al. 2022).
In summary, the fact that the microbiomes of the algal cultures and the sponge host microbiomes are so similar, sharing a high proportion of their taxa, strongly points to a crucial role of the prokaryotic symbionts in the growth and metabolic stability of both the algae and sponges. Freshwater sponges and their algal partners offer many opportunities to examine important questions in host:symbiont interactions from molecular, genetic, cellular, developmental, ecological, and evolutionary perspectives.
Data Availability
All data is provided in figures and supplemental.
References
Agnes B, Brøndsted HV (1953) The effect of symbiotic zoochlorellae on the germination rate of gemmules of Spongilla lacustris (L.). Viddensk Medd Dan Naturhist Foren 115:133–144
Annenkova NV, Lavrov DV, Belikov SI (2011) Dinoflagellates associated with freshwater sponges from the ancient Lake Baikal. Protist 162:222–236. https://doi.org/10.1016/j.protis.2010.07.002
Apprill A, McNally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:1290137. https://doi.org/10.3354/ame01753
Arillo A, Bavestrello G, Burlando B, Sarà M (1993) Metabolic integration between symbiotic cyanobacteria and sponges: a possible mechanism. Mar Biol 117:159–162. https://doi.org/10.1007/BF00346438
Astafyeva Y, Gurschke M, Qi M, Bergmann L, Indenbirken D, de Grahl I, Katzowitsch E, Reumann S, Hanelt D, Alawi M, Streit WR, Krohna I (2022) Microalgae and Bacteria Interaction—Evidence for Division of Diligence in the Alga Microbiota. Microbiol Spectr 4:e00633-e722. https://doi.org/10.1128/spectrum.00633-22
Bathia J, Schröder K, Fraune S, Lachnit T, Rosenstie P, Bosch TCG (2022) Symbiotic Algae of Hydra viridissima Play a Key Role in Maintaining Homeostatic Bacterial Colonization. Front Microbiol 13:869666. https://doi.org/10.3389/fmicb.2022.869666
Berner T, Titlyanov E (1992) Are zoochlorellae from Lake Baikal sponges shade adapted? Symbiosis 14:465–473
Bil K, Titlyanov E, Berner T, Fomina I, Muscatine L (1999) Some aspects of the physiology and biochemistry of Lubomirska baikalensis, a sponge from Lake Baikal containing symbiotic algae. Symbiosis 26:179–191
Bock C, Pröschold T, Krienitz L (2010) Two new Dictyosphaerium-morphotype lineages of the Chlorellaceae (Trebouxiophyceae): Heynigia gen. nov. and Hindakia gen. nov. Eur J Phycol 45:267–277. https://doi.org/10.1080/09670262.2010.487920
Brandt K (1882) Über das Zusammenleben von Algen und Thieren. Biol Centralbl 1:524–527
Burja AM, Tamagnini P, Bustard MT, Wright PC (2001) Identification of the green alga, Chlorella vulgaris (SDC1) using cyanobacteria derived 16S rDNA primers: targeting the chloroplast. Fems Microbiol Lett 202:195–203. https://doi.org/10.1111/j.1574-6968.2001.tb10803.x
Castro-Rodriguez G (1930) De la symbiose entre la Spongilla lacustris et les Zoochlorelles. Contribution ál’étude de la nutrition des spongiaires. Remarques sur la fécondation des spongiaires. Ann Soc Roy Zool Belgique 61:113–121
Cerrano C, Calcinai B, Cucchiari E, Di Camillo C, Totti C, Bavestrello G (2004) The diversity of relationships between Antarctic sponges and diatoms: the case of Mycale acerata (Porifera, Demospongiae). Polar Biol 27:231–237. https://doi.org/10.1007/s00300-003-0581-1
Cirri E, Pohnert G (2019) Algae−bacteria interactions that balance the planktonic microbiome. New Phytol 223:100–106. https://doi.org/10.1111/nph.15765
Cook CB (1983) Metabolic interchange in algae-invertebrate symbiosis. Internat Rev Cytol Suppl 14:177–210
Colgren J, Nichols SA (2022) MRTF specifies a muscle-like contractile module in Porifera. Nat Commun 13:4134. https://doi.org/10.1038/s41467-022-31756-9
Chernogor L, Denikina N, Kondratov I, Solovarov I, Khanaev I, Belikov S, Ehrlich H (2013) Isolation and identification of the microalgal symbiont from primmorphs of the endemic freshwater sponge Lubomirskia baicalensis (Lubomirskiidae, Porifera). Eur J Phycol 48:497–508. https://doi.org/10.1080/09670262.2013.862306
Darienko T, Pröschold T (2015) Genetic variability and taxonomic revision of the genus Auxenochlorella (Shihira et Krauss) Kalina et Puncocharova (Trebouxiophyceae, Chlorophyta). J Phycol 51:394–400. https://doi.org/10.1111/jpy.12279
Díez-Vives C, Koutsouveli V, Conejero M, Riesgo A (2022) Global patterns in symbiont selection and transmission strategies in sponges. Front Ecol Evol 10. https://doi.org/10.3389/fevo.2022.1015592
Elliott GRD, Leys SP (2010) Evidence for Glutamate, GABA and NO in coordinating behaviour in the sponge Ephydatia muelleri (Demospongiae, Spongillidae). J Exp Biol 213(13):2310–2321. https://doi.org/10.1242/jeb.039859
Elliott GRD, Leys SP (2007) Coordinated behaviour in a freshwater sponge: orchestrated contractions effectively expel water from the aquiferous system. J Exp Biol 210:3736–3748. https://doi.org/10.1242/jeb.003392
Ereskovsky A, Chernogor L, Belikov S (2016) Ultrastructural description of development and cell composition of primmorphs in the endemic Baikal sponge Lubomirskia baicalensis. Zoomorphology 135:1–17. https://doi.org/10.1007/s00435-015-0289-0
Ereskovsky A, Rinkevich B, Somorjai I (2022) Adult stem cells host intracellular symbionts: The poriferan archetype. In: Balarin L, Rinkevich B, Hobmayer B (eds) Advances in aquatic invertebrate stem cell research. MDPI Books. hal-03592793. https://doi.org/10.3390/books978-3-0365-1635-6-2
Fama P, Wysor B, Kooistra W, Zuccarello GC (2002) Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. J Phycol 38:1040–1050. https://doi.org/10.1046/j.1529-8817.2002.t01-1-01237.x
Frost T, Williamson C (1980) In situ determination of the effect of symbiotic algae on the growth of the freshwater sponge Spongilla lacustris. Ecol 61:1361–1370. https://doi.org/10.2307/1939045
Fuentes JL, Garbayo I, Cuaresma M, Montero Z, González-del-Valle M, Vílchez C (2016) Impact of Microalgae-Bacteria Interactions on the Production of Algal Biomass and Associated Compounds. Mar Drugs 14:100. https://doi.org/10.3390/md14050100
Gaikwad S, Shouche YS, Gade WN (2016) Microbial community structure of two freshwater sponges using Illumina MiSeq sequencing revealed high microbial diversity. AMB Express 6:40. https://doi.org/10.1186/s13568-016-0211-2
Gaino E, Manconi R, Pronzato R (1995) Organizational plasticity as a successful conservative tactics in sponges. Anim Biol 4:31–43
Geraghty S, Koutsouveli V, Hall C, Chang L, Sacristan-Soriano O, Hill MS, Riesgo A, Hill AL (2021) Establishment of Host–Algal Endosymbioses: Genetic Response to Symbiont Versus Prey in a Sponge Host. Genome Biol Evol 13:evab252. https://doi.org/10.1093/gbe/evab252
Gilbert JJ, Allen HL (1973) Chlorophyll and primary productivity of some green, freshwater sponges. Int Rev Ges Hydrobiol 58:633–658
Haberkorn I, Walser J-C, Helisch H, Böcker L, Belz S, Schuppler M, Fasoulas S, Mathys A (2020) Characterization of Chlorella vulgaris (Trebouxiophyceae) associated microbial communities. J Phycol 56:1308–1322. https://doi.org/10.1111/jpy.13026
Hall C, Camilli S, Dwaah H, Kornegay B, Lacy C, Hill MS, Hill AL (2021) Freshwater sponge hosts and their green algae symbionts: a tractable model to understand intracellular symbiosis. PeerJ 9:e10654. https://doi.org/10.7717/peerj.10654
Hall C, Rodriguez M, Garcia J, Posfai D, Dumez R, Wictor E, Quintero O, Hill MS, Rivera A, Hill AL (2019) Secreted frizzled related protein is a target of PaxB and plays a role in aquiferous system development in the freshwater sponge, Ephydatia muelleri. PLoS One 14:e0212005. https://doi.org/10.1371/journal.pone.0212005
Handa S, Nakahara M, Tsubota H, Deguchi H, Masuda Y, Nakano T (2006) Choricystis minor (Trebouxiophyceae, Chlorophyta) as a symbiont of several species of freshwater sponge. Hikobia 14:365–373. https://doi.org/10.1007/s13199-020-00711-x
Hill MS (1996) Symbiotic zooxanthellae enhance boring and growth rates of the tropical sponge Anthosigmella varians forma varians. Mar Biol 125:649–654. https://doi.org/10.1007/BF00349246
Hill A, Nguyen I, Hill M (2020) Isolation of green algal symbionts from freshwater sponges and subsequent reinfection of sponge tissues. protocols.io. https://doi.org/10.17504/protocols.io.bmuzk6x6
Hudspith M, de Goeij JM, Streekstra M et al (2022) Harnessing solar power: photoautotrophy supplements the diet of a low-light dwelling sponge. ISME J. https://doi.org/10.1038/s41396-022-01254-3
Kalyaanamoorthy S, Minh BQ, Wong TKF, van Haeleler A, Jermiin LS (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. https://doi.org/10.1038/nmeth.4285
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kenny NJ, Plese B, Riesgo A, Itskovich VB (2019) Symbiosis, selection, and novelty: freshwater adaptation in the unique sponges of Lake Baikal. Mol Biol Evol 36:2462–2480
Kenny NJ, Francis WR, Rivera-Vicéns RE et al (2020) Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat Commun 11:3676. https://doi.org/10.1038/s41467-020-17397-w
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120
Krohn-Molt I, Alawi M, Förstner KU, Wiegandt A, Burkhardt L, Idenbirken D, Theiβ M, Grundhoff A, Kehr J, Tholey A, Streit WR (2017) Insights into Microalga and Bacteria Interactions of Selected Phycosphere Biofilms Using Metagenomic, Transcriptomic, and Proteomic Approaches. Front Microbiol 8:1941. https://doi.org/10.3389/fmicb.2017.01941
Kulakova NV, Denikina NN, Belikov S (2014) Diversity of Bacterial Photosymbionts in Lubomirskiidae Sponges from Lake Baikal. Int J Biodivers. 152097. https://doi.org/10.1155/2014/152097
Kulakova N, Kashin S, Bukin Y (2020) The genetic diversity and phylogeny of green microalgae in the genus Choricystis (Trebouxiophyceae, Chlorophyta) in Lake Baikal. Limnol 21:15–24. https://doi.org/10.1007/s10201-019-00587-x
Lewin RA (1966) Kultivo de Zoochlorella apartigita el spongo. Scienca Revuo 17:33–37
Leys SP, Grombacher L, Hill A (2019) Hatching and freezing gemmules from the freshwater sponge Ephydatia muelleri. protocols.io. https://doi.org/10.17504/protocols.io.863hzgn
Ludeman DA, Farrar N, Riesgo A, Paps J, Leys SP (2014) Evolutionary origins of sensation in metazoans: functional evidence for a new sensory organ in sponges. BMC Evol Biol 14(3). https://doi.org/10.1186/10.1186/1471-2148-14-3.
Marin B, Klingberg M, Melkonian M (1998) Phylogenetic relationships among the cryptophyta: analyses of nuclear-encoded SSU rRNA sequences support the monophyly of extant plastid-containing lineages. Protist 149:265–276. https://doi.org/10.1016/S1434-4610(98)70033-1
Marin B, Palm A, Klingberg M, Melkonian M (2003) Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA Sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 154:99–145. https://doi.org/10.1078/143446103764928521
Masuda Y (1990) Electron microscopic study on the zoochlorellae of some freshwater sponges. In: Klais Rützler (ed.) New Perspectives in Sponge Biology. Smithsonian Institution Press Washington, Washington, DC, pp 467–71
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. https://doi.org/10.1093/molbev/msaa015
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast Approximation for Phylogenetic Bootstrap. Mol Biol Evol 30:1188–1195. https://doi.org/10.1093/molbev/mst024
Mitchell J, Nichols S (2019) Diverse cell junctions with unique molecular composition in tissues of a sponge (Porifera). EvoDevo 10:26. https://doi.org/10.1186/s13227-019-0139-0
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, and Wagner H (2018) vegan: Community Ecology Package. Ordination methods, diversity analysis and other functions for community and vegetation ecologists. Version 2.5–1. https://CRAN.R-project.org/package=vegan
Okuda N, Yamamoto A, Satoh Y, Fujimoto Y, Kamishima Y (2002) Role of Symbiotic Algae on Gemmule Germination of a Freshwater Sponge, Radiospongilla cerebellata. Chugokugakuen J 1:7–12
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. https://doi.org/10.1111/1462-2920.13023
Pita L, Rix L, Slaby BM, Franke A, Hentschel U (2018) The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6:46. https://doi.org/10.1186/s40168-018-0428-1
Pröschold T, Darienko T (2020) Choricystis and Lewiniosphaera gen. Nov. (Trebouxiophyceae Chlorophyta), two different green algal endosymbionts in freshwater sponges. Symbiosis 82:175–188. https://doi.org/10.1007/s13199-020-00711-x
Pröschold T, Darienko T, Silva PC, Reisser W, Krienitz L (2011) The systematics of "Zoochlorella" revisited employing an integrative approach. Environ Microbiol 13:350–364. https://doi.org/10.1111/j.1462-2920.2010.02333.x
Ramanan R, Kim B-H, Cho D-H, Oh H-M, Kim H-S (2016) Algae-bacteria interactions: Evolution, ecology and emerging interactions. Biotechnol Adv 34:14–29. https://doi.org/10.1016/j.biotechadv.2015.12.003
Ramsby BD, Hill MS, Thornhill DJ, Steenhuizen SF, Achlatis M, Lewis AM, LaJeunesse TC (2017) Sibling species of mutualistic Symbiodinium Clade G from bioeroding sponges in the Western Pacific and Western Atlantic oceans. J Phycol 53(5):951–960. https://doi.org/10.1111/jpy.12576
Rasmont R (1970) Some new aspects of the physiology of freshwater sponges. In: Fry WG (ed) The Biology of thePorifera. Academic Press, London, pp 415–422
Reisser W (1984) The taxonomy of green algae endosymbiotic in ciliates and a sponge. Br Phycol J 19:309–318. https://doi.org/10.1080/00071618400650361
Rützler K (1985) Associations between Caribbean sponges and photosynthetic organisms. New perspectives in sponge biology. In: Rützler K (ed). Smithsonian Institution Press, Washington, DC, 455-466
Saller U (1989) Microscopical aspects on symbiosis of Spongilla lacustris (Porifera, Spongillidae) and green algae. Zoomorphology 108:291–296. https://doi.org/10.1007/BF00312161
Saller U (1991) Symbiosis of Spongilla lacustris (Spongilledae) and green algae. Algae uptake, distribution and final whereabouts. In: Reitner J, Keupp H (eds) Fossil and Sponges. Springer-Verlag, Berlin, pp 299–305
Sand-Jensen K, Pedersen MF (1994) Photosynthesis by symbiotic algae in the freshwater sponges, Spongilla lacustris. Limnol Oceanogr 39:551–561. https://doi.org/10.4319/lo.1994.39.3.0551
Schenkelaars Q, Quintero O, Hall C, Fierro-Constain L, Renard E, Borchiellini C, Hill AL (2016) ROCK inhibition abolishes the establishment of the aquiferous system in Ephydatia muelleri (Porifera, Demospongiae). Dev Biol 412:298–310. https://doi.org/10.1016/j.ydbio.2016.02.026
Scott FJ, Wetherbee R, Kraft GT (1984) The morphology and development of some prominently stalked southern Australian Halymeniaceae (Cryptonemiales, Rhodophyta). II. The sponge-associated genera Thamnoclonium Kuetzing and Codiophyllum Gray. J Phycol 20:286–295. https://doi.org/10.1111/j.0022-3646.1984.00286.x
Simpson T (1984) The Cell Biology of Sponges. Springer, NewYork, p.678
Sun X, Xiao-Wei WU, Xing-Wen LI, Pei LQ (2009) Molecular identification of Chlorella strains based on sequence analysis of nuclear rDNA ITS and chloroplast rbcL gene. J Fish China 33:565–571
Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M (2014) Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS One 9(8):e105592. https://doi.org/10.1371/journal.pone.0105592
Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol Mol Biol R 71:295–347. https://doi.org/10.1128/MMBR.00040-06
Thacker R, Freeman CJ (2012) Sponge-microbe symbioses: Recent advances and new directions. Adv Mar Biol 62:57–111. https://doi.org/10.1016/B978-0-12-394283-8.00002-3
Thomas T, Moitinho-Silva L, Lurgi M, BjoÈrk JR et al (2016) Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 7:11870. https://doi.org/10.1038/ncomms11870
Tronchin E, Samaai T, Anderson RJ, Bolton JJ (2006) Sponge–seaweed associations in species of Ptilophora (Gelidiaceae, Rhodophyta). Phycol Res 54:140–148. https://doi.org/10.1111/j.1440-1835.2006.00422.x
Webster NS, Taylor MW (2012) Marine sponges and their microbial symbionts: Love and other relationships. Environ Microbiol 14:335–346. https://doi.org/10.1111/j.1462-2920.2011.02460.x
Weisz JB, Massaro AJ, Ramsby BD, Hill MS (2010) Zooxanthellar symbionts shape host sponge trophic status through translocation of carbon. Biol Bull 219:189–197. https://doi.org/10.1086/BBLv219n3p189
Wilkinson CR, Fay P (1979) Nitrogen fixation in coral reef sponges with symbiotic cyanobacteria. Nature 279:527–529. https://doi.org/10.1038/279527a0
Wilkinson CR (1980) Nutrient translocation from green algal symbionts to the freshwater sponge Ephydatia fluviatilis. Hydrobiologia 75:241–250
Wilkinson CR (1983) Net primary productivity in coral reef sponges. Science 219:410–412. https://doi.org/10.1126/science.219.4583.410
Williamson CE (1979) An ultrastructural investigation of algal symbiosis in white and green Spongilla lacustris (L.) (Porifera: Spongillidae). Trans Am Microsc Soc 98:59–77
Windsor-Reid P, Matveev E, McClymont A, Posfai D, Hill AL, Leys SP (2018) Wnt signaling and polarity in freshwater sponges. BMC Evol Biol 18:12. https://doi.org/10.1186/s12862-018-1118-0
Windsor PJ, Leys SP (2010) Wnt signaling and induction in the sponge aquiferous system: evidence for an ancient origin of the organizer. Evol Dev 12:481–490. https://doi.org/10.1111/j.1525-142X.2010.00434.x
Zhang F, Blasiak LC, Karolin JO, Powell RJ, Geddes CD, Hill RT (2015) Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges. Proc Natl Acad Sci U S A 11214:4381–4386. https://doi.org/10.1073/pnas.1423768112
Zou S, Fei C, Wang C, Gao Z, Bao Y, He M, Wang C (2016) How DNA barcoding can be more effective in microalgae identification: a case of cryptic diversity revelation in Scenedesmus (Chlorophyceae). Sci Rep 6:36822. https://doi.org/10.1038/srep36822
Acknowledgements
This work was supported by a grant from the Gordon and Betty Moore Foundation (#9332) and by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (#P20GM103423). CD-V received funding from a fellowship from “la Caixa” Foundation (ID 100010434), code LCF/BQ/PI22/11910040.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Katelyn Hustus Cristina Díez-Vives co-first authors.
Supplementary Information
Below is the link to the electronic supplementary material.
13199_2023_934_MOESM1_ESM.pdf
Fig. S1 Molecular phylogeny of Chlorellaceae based on the chloroplast marker gene tufA. The tree is inferred by maximum likelihood and the nodal supports are ultrafast bootstraps. The algal isolates from this study are in bold. Note that only bootstrap values over 70 are shown. (PDF 3102 KB)
13199_2023_934_MOESM2_ESM.pdf
Fig. S2 Molecular phylogeny of Chlorellaceae based on the chloroplast marker rbcL. The tree is inferred by maximum likelihood and the nodal supports are ultrafast bootstraps. The algal isolates from this study are in bold. Note that only bootstrap values over 70 are shown. (PDF 2157 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hustus, K., Díez-Vives, C., Mitsi, K. et al. Algal symbionts of the freshwater sponge Ephydatia muelleri. Symbiosis 90, 259–273 (2023). https://doi.org/10.1007/s13199-023-00934-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-023-00934-8