Abstract
Animals, plants, and fungi live in a microbe-dominated world. Investigating the interactions and processes at the host-microbe interface offers insight to how bacteria influence the development, health, and disease of the host. Optimization of existing imaging technologies and development of novel instrumentation will provide the tools needed to fully understand the dynamic relationship that occurs at the host-microbe interface throughout the lifetime of the host. In this review, we describe the current methods used in cryo-electron microscopy (cryo-EM) including cryo-fixation, sample processing, FIB-SEM, and cryotomography. Further, we highlight the new advances associated with these methods that open the cryo-EM discipline to large, complex multicellular samples, like symbiotic tissues. We describe the advantages and challenges associated with correlative imaging techniques and sample thinning methods like lift-out. By highlighting recent pioneering studies in the large-volume or symbiotic sample workflows, we provide insight into how symbiotic model systems will benefit from cryo-EM methods to provide artefact-free, near-native, macromolecular-scale resolution imaging at the host-microbe interface throughout the development and maintenance of symbiosis. Cryo-EM methods have brought a deep fundamental understanding of prokaryotic biology since its conception. We propose the application of existing and novel cryo-EM techniques to symbiotic systems is the logical next step that will bring an even greater understanding how microbes interact with their host tissues.
Similar content being viewed by others
Biologists have gained an increasing awareness of the role of microbes in the overall health and disease of humans, as well as other animals, plants, and fungi. Animals and plants do not live in isolation, but rather build intimate relationships with a myriad of microbes (Gilbert et al. 2012; McFall-Ngai et al. 2013; Fronk & Sachs 2022). As such, symbiotic research is a rapidly growing research field. Through multidisciplinary and multiscale approaches, we continue to gain a better understanding of the impact of host-microbe interactions. They can form transient or chronic associations with microbial species throughout the lifetime of the organism, which determine the health, development, metabolism, and behavior of the host (McFall-Ngai et al. 2013). With the use of different model systems and integrative techniques including for example proteomics, transcriptomics, advanced imaging methods, the research community is continuing to gain deeper insight into the interactions at the interface between microbe and host tissue.
Of particular interest are the single cell interactions between host and microbe at different stages of life of the host. The research can be broken down into different categories, including identifying the biogeographical location of the symbiont, which host cell(s) it is in contact with, which microstructures and molecules are used to interact and communicate between host and symbiont, and how this relationship is maintained through health and disease.
This review focuses on the contributions and future directions of advanced imaging methods to the greater understanding of the symbiotic relationships between animal and microbe; in particular, cryo-electron microscopy (cryo-EM) techniques and workflows. Histological tissue preparations and confocal imaging techniques have been, and will remain, valuable to understand the interactions of microbes with host tissue on a greater scale; these methods remain affordable, rapid, and informative (Aulner et al. 2018; López-Jiménez & Mostowy 2021). Microbes, specifically bacteria, interact with host tissue using a wide variety of extracellular attachment structures. For example, surface pili are understood to be an essential component for host cell adhesion and colonization of symbiotic tissues (Sharma et al. 2021). Flagella are a well-recognized virulence factor for pathogenic bacteria. Additionally, recent work implies a role of the flagella in developing and maintaining a healthy gut microbiota (Akahoshi & Bevins 2022). Furthermore, bacteria have several different classes of secretion systems. The role these systems play in the development and maintenance of symbiosis is not yet fully explored (Cao et al. 2018; Jani & Cotter 2010; Lin et al. 2021; Park et al. 2018; Russell et al. 2014).
To gain insight into the fine-scale interactions between host and microbes, the highest resolution imaging techniques like electron microscopy (EM) are required. The magnification power of transmission electron microscopy (TEM) allows the visualization of macromolecular structures between and inside cells inside tissues. For conventional room temperature sample preparation, the tissue undergoes extensive processing steps, including chemical fixation, dehydration, heavy metal staining, plastic embedding, and sectioning prior to imaging in an electron microscope. These sample preparation methods can introduce artefacts and obscure fine structural details, resulting in non-native structural and conformational changes (Ayache et al. 2019).
1 Cryo-electron tomography
Cryo-electron tomography (cryo-ET) is a relatively new field of electron microscopy that bypasses the challenges faced with chemically fixed samples (Milne & Subramaniam 2009). It provides the means to acquire three-dimensional images of the sample in a near native state and at macromolecular resolution. The limit of resolution for cryo-ET is constantly being pushed by optimizing sample preparation and data processing methods. Attainable resolution is highly sample dependent. In ideal cases, sub-nanometer resolution has been achieved (Turk & Baumeister 2020), but the resolution of thicker samples such as whole bacterial cells is typically in the range of 2–4 nm.
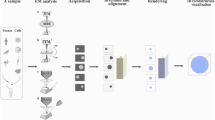
To prepare a sample suitable for cryo-ET, cells are grown on, or applied to, an EM support grid, then flash frozen in a liquid-nitrogen cooled cryogen (Fig. 1) (Dobro et al. 2010). This plunge-freezing is rapid enough to prevent the formation of ice crystals and transforms the sample into a glass-like state by a process called vitrification. This method preserves the ultrastructure of the sample without the typical artefacts associated with traditional EM.

Adapted from Turk & Baumeister 2020
Overview of currently developed cryo-ET methods. First row illustrates the range of sizes and diversity of samples used in cryo-ET studies. The second row illustrates the vitrification method suitable for each class of sample type or size. Third row shows a visual representation of how larger samples are manipulated to a suitable size for cryo-ET, and then also how cryo-ET data is collected from sufficiently frozen and thinned samples.
The vitrified samples can be directly imaged using a cryo-transmission electron microscope (cryo-TEM). For cryo-ET, images are collected while rotating the sample in the microscope relative to the electron beam, resulting in a tilt series of images for each target. Computationally, the tilt series is then used to generate a high-resolution three-dimensional volume of the target called a tomogram (Harapin et al. 2015; Milne & Subramaniam 2009; Oikonomou & Jensen 2017).
2 Sample vitrification
At first, cryo-ET workflows were limited to very thin samples such as suspensions of viruses, isolated organelles, or prokaryotic cells. This is due to two limiting factors: first, thicker or more complex samples are not completely vitrified using plunge-freezing methods, which can freeze up to a maximum sample thickness of 5 µm (Bouchet-Marquis & Hoenger 2011). Additionally, the electron beam must penetrate the entire sample to generate an image. Therefore, samples that exceed 0.5 -1 µm are unsuitable for cryo-ET. Despite these limitations, this method has proven to be a very powerful tool to unravel structure and function of cellular structures. For example, bacterial molecular machines have been imaged at high resolution using cryo-ET, including chemotaxis arrays, flagellar motor, secretion systems, ribosomes and more (Liedtke et al. 2022).
In recent years, significant efforts have been made to overcome the size limitations of the samples that can be imaged with cryo-ET. In order to enable the proper vitrification of thicker samples, high-pressure can be applied during the freezing process, which prevents sample expansion due to crystallization (Fig. 1) (Moor 1987; D. Studer et al. 2001). This allows complete vitrification of samples up to 200 µm (Daniel Studer et al. 2008). For high-pressure freezing, the biological sample is loaded into copper or aluminum planchette carriers with the addition of a cryoprotectant. Subsequently, the sample is frozen with 2000 atm pressure in milliseconds. If the sample is below the size limitation of 200 µm in depth, the biological sample is frozen within a block of ice inside the planchette. The sample is then ready for further processing. Almost any sample is suitable for high pressure freezing (HPF), unless there are gaseous spaces like vacuoles inside the tissue, which can implode during HPF, resulting in gross tissue malformations. Thus, HPF provides the means for cryo-fixation of larger biological samples. It preserves their near-native structural state without any chemical-fixation related artefacts. The maximum size for HPF samples is limited by the physical properties and rate of heat transfer of water under elevated pressure.
Larger samples like tissue extractions, larvae, organelles, and cell suspensions can now easily be frozen and processed for cryo-EM, which expanded the application of highest-resolution imaging methods once limited to single cell preparation to tissues, organelles, and complex multicellular systems.
3 Sample thinning
The second limitation that needs to be overcome is the thickness of the samples that can be imaged. To enable imaging of thicker samples that can be vitrified by HPF, they must be thinned before cryo-EM imaging (Fig. 1). Several methods are now available to do this, such as trimming and sectioning, or focused ion beam-milling (FIB-milling), or a combination of all three. Cryo-ultramicrotomes (CUMT) are used with diamond knives to trim and/or section HPF samples to within the size requirements for downstream processing or imaging. Samples can be trimmed and sectioned into ultrathin sections directly suitable for cryo-EM with a method called cryo-EM of vitreous sections, or CEMOVIS. CEMOVIS is technically challenging and requires optimization of cutting speed, section thickness, and handling techniques. First, the frozen sample is trimmed with a diamond knife into a pyramid with a flat square top. Then, a diamond knife cuts sections from the prepared flat top which are guided onto a mesh support grid (Al-Amoudi et al. 2004). The notable disadvantages of CEMOVIS concern artefacts derived from mechanical stress during cutting with the knife. Compression, crevasses, curtaining, and knife marks in micrographs are the most common artefacts caused by the knife, which can distort molecular details (Al-Amoudi et al. 2005).
Another established method for sample thinning is called focused ion beam milling scanning electron microscopy, or FIB-SEM. A SEM beam is combined into one machine with a gallium-ion beam which is used to mill away tissue at precisely targeted regions of interest with the goal to create thinned areas suitable for cryo-ET. A support grid with a frozen sample is simultaneously monitored with the SEM beam, while the gallium-ion beam is used to mill away at the sample at a predetermined angle. A typical milling pattern creates a thin window of tissue, or lamella, between 100–300 nm in thickness. Operation of the FIB-SEM is comparatively straightforward, where the bulk of the work is involved in optimizing the shape of the milling window for lamellae formation, angle and thickness of lamellae, SEM beam conditions, and gallium-ion beam conditions (Hayles and Winters 2021). While lamellae are delicate and remain a challenge to transfer to the final imaging cryo-TEM without breaking, FIB-SEM overcomes a lot of the disadvantages of processing samples with the CUMT (Hayles & de Winter 2021).
The specialized nature of these instruments, the level of training required to process samples and data, and the physical limitations in sample size are a few reasons why the application of cryo-EM methods has previously been restricted to only the most well-studied cellular models or isolated structures. However, innovative technological advances now offer the chance for larger, more complex systems to take advantage of highest possible resolution imaging methods of near-native samples. Symbiosis model systems are a logical choice to apply these large volume cryo-EM methods. Symbiotic host tissues interact with microbes using nanostructures, which require high resolution imaging to gain insight into structure and function of these interactions. Bringing these samples to the cryo-EM discipline provides a great opportunity to see the host-microbe interface at unprecedented resolution, which we expect will become an essential tool for microbiome research in the future. Here we present notable examples where novel cryo-EM techniques have successfully been combined with established cryo-EM workflows to study larger, complex samples. Layering together these processes give us a window into the spatial and temporal interactions of hosts with their microbes. These pioneering studies have set the foundation for how these technologies and methods can be applied directly to study complex host-microbe interactions.
4 Single cell host symbiosis and cryo-ET
In host-microbe associations, the host symbiotic tissue or organism is typically much larger than the microbe, usually far-exceeding the size limitation for conventional cryo-fixation methods like plunge freezing or even high pressure freezing. Therefore, such samples were not suitable for cryo-EM imaging methods. A notable exception is a system with a single-celled amoebae, Acanthamoeba castellanii (the host) and environmental Chlamydiae (the microbe). While the majority of work on Chlamydia spp. is related to its function as a human and animal pathogen, some studies have been investigating environmental chlamydiae (Collingro et al. 2020). These cultured environmental chlamydiae are strictly intracellular and use amoebae as hosts, where the internalization process of the microbe and host-organelle recruitment is mediated by a dynamic bacterial structure called the type-3 secretion system (T3SS) (Pilhofer et al. 2014). Much of the foundation of Chlamydiae biology was dependent on chemical fixation, dehydration, plastic embedding, and heavy metal staining which are known to cause membrane artefacts, potential misrepresentation of structures, or loss of entire cellular components (Pilhofer et al. 2010).
In 2014, Pilhofer et al. imaged bacteria inside their host cells for the first time, using cryo-fixation. Three environmental strains of Chlamydiae and their amoebae host were high-pressure frozen, followed by the CEMOVIS method (Pilhofer et al. 2014). Results from their work and another group’s work (Nans et al. 2014) show localization of bacteria within host cells, the surrounding membrane morphology, and T3SS needle structures engaging with host cell membranes (Figs. 2a-2d). In addition, micrographs from this work provide evidence that a previously thought-to-be infectious life stage, the crescent body, is an artefact derived from chemical fixation. Additionally, conventional EM methods have been implicated with mischaracterizing nucleoid and periplasmic membrane structures, a result from harsh sample preparation methods (Pilhofer et al. 2010). This is the first example of a study that successfully imaged bacteria inside their host at near-native, frozen-hydrated state using cryo-EM methods which avoid and uncover artefacts. These studies provided new insights into the biology of the host-bacterial interface. In 2018, another noteworthy example of host-microbe visualization was successfully accomplished using a Salmonella-HeLa cell symbiotic system (Park et al. 2018). This group was able to show the bacterial nanostructure, the T3SS, interacting with cultured HeLa cell plasma membranes and follow up with high-resolution structural analyses (Figs. 2e and 2f). These studies are great examples of how cryo-ET methods can provide additional insights into well-studied structures and further our understanding of complex interactions at the host-microbe interface.
Bacterial secretion systems at the host cell interface. Cryotomogram slice showing Simkania type 3 secretion system (T3SS) needle (arrowhead) interacting with host cells (a), enlarged in (b), adapted from Pilhofer et al. 2014. Cryotomogram slice showing four Chlamydia cells interacting with cultured host cell (c). White boxes outline T3SS directly contacting host cell. Three-dimensional surface representation of host surface (yellow), bacterial cells (green/blue), and T3SS (red), generated from segmentation (d), (c) and (d) adapted from Nans et al. 2014. Cryotomogram slice showing Salmonella typhimurium mini cell interacting with HeLa host cell via T3SS (e), plasma membrane (PM), outer membrane (OM), and inner membrane (IM). Three-dimensional surface representation of S. typhimurium’s (green) T3SS (blue spike) interacting with host PM (red) with underlying actin filaments (orange) (f), (e) and (f) adapted from Park et al. 2018
5 Multicellular organism sample preparation and cryo-ET
The majority of symbiotic systems involve larger, multicellular hosts which face additional challenges or limitations in cryo-EM workflows as compared to single cell systems. Due to their size, symbiotic multicellular tissues cannot be successfully frozen using a plunge freezer and therefore require specialized equipment like the HPF for cryo-fixation. While each sample type requires its own optimization for HPF methods (Mcdonald et al. 2007), as long as the sample is within the size limitations of HPF vitrification, studying larger samples with cryo-EM is now possible. Second, once the sample is frozen it must be reduced in size to be suitable for cryo-ET. As outlined above, methods for sample thinning include CEMOVIS (sections) or FIB-SEM (lamellae). Creating lamellae from large multicellular samples often requires unrealistic milling times and comes with the added challenge of target localization within sample before, during and after milling. Once lamellae are created, sample handling and transfer between machines continues to provide challenges regarding stability and ice contamination. These workflows are difficult, low-throughput, and required a high level of technical training.
Shortly after the single-cell host work with Chlamydiae-amoebae was reported, an innovative workflow was developed to optimize sample preparation and cryo-ET of multicellular samples, specifically Caenorhabditis elegans embryos and worms (Harapin et al. 2015; Mahamid et al. 2015). A whole C. elegans embryo was frozen by HPF, thinned using FIB-SEM, micromanipulated using a novel technique called lift-out, and imaged using cryo-ET. Subsequent work led to the development of novel and improvement on existing correlative imaging instruments to increase throughput and aide in accuracy during sample preparation workflows (Gorelick et al. 2019). Lift-out is a technique that follows successful lamellae milling. A micromanipulator within the column grasps the thinned lamellae, removes it from surrounding substrate, and transfers it to a support grid, which gets transferred into the cryo-TEM (Figs. 3a and 3b). The use of the lift-out method reduces the milling time required in thicker samples. A single lamellae can take an hour to mill on thin samples, but can take 10 s of hours on thicker multicellular samples (Harapin et al. 2015). Lift-out streamlines the sample preparation process by no longer requiring the removal of all the tissue above and below the lamellae, but just enough for the pinchers to grasp the lamellae. The relative location of the region of interest within the sample will be critical to the successful application of FIB-SEM lift-out on a sample, i.e., the region of interest will need to be close enough to the surface of the sample to be accessible by both the gallium beam and the lift-out pinchers. Optimizing orientation of sample during freezing followed by sufficient trimming of the sample prior to lamellae formation will ensure a lamella can be milled where the host-microbe interface is found. Lift-out techniques are promising for the future of FIB-milling but are technically challenging and require specialized instruments. Alternatively, a newly described FIB-milling technique called the Waffle Method has been developed that does not require the specialized equipment needed in lift out methods (Kelley et al. 2022). The Waffle Method is an approach in which the sample is high pressure frozen directly onto a mesh grid. The FIB-SEM is used to mill grid lines into the top of the frozen sample, into a ‘waffle pattern’, as a sort of atlas. Two trenches are milled within each grid square, then an angled lamella is milled between the trenches. Their results show this method to increase throughput and stability and is applicable to many different sample types. We expect ongoing development of creative solutions like the Waffle Method to address the challenges seen in the large-volume sample processing.
Advanced large-volume techniques with FIB-SEM. Attachment of micromanipulator to milled lamellae for lift-out (a), outlined yellow region shows remaining attachment point to be FIB-milled away. Excised lamellae attached to micromanipulator (b), (a) and (b) adapted from Parmenter & Nizamudeen 2021. Correlative microscopy images of C. elegans worm using fluorescence and SEM with PIE-scope before (c), and after milling a lamella (d), (c) and (d) adapted from Gorelick et al. 2019
During lamellae preparation, accurate target localization in the region of interest is essential. However, to mill lamellae containing features of the internal host-microbe interface or otherwise heterogeneously distributed features, additional imaging modes are required. Correlative microscopes combine the power of epifluorescence light microscopy on an cryo-EM sample, like the Cryo-CLEM (Liedtke et al. 2022). However, this is a separate instrument which then brings the risk of ice contamination and sample loss during each additional cryo-transfer. In 2019, the Photon Ion Electron Microscope, or the PIE-scope, was designed to overcome these challenges by integrating a light microscopy objective inside the FIB-SEM column, facilitating real-time verification of fluoro-labelled targets in lamellae during milling (Gorelick et al. 2019) (Figs. 3c and 3d). We expect this to be particularly useful for symbiotic tissues, i.e., a sample containing GFP-labelled symbionts which would aide in rapidly and accurately determining milling regions.
Method development and workflow optimization remains a high priority focus for processing large volumes samples for cryo-ET studies. These workflows have technical challenges and are often time consuming. Recent unpublished work has taken a slightly different approach to overcome the size limitations of traditional plunge freezing methods (Bäuerlein et al. 2021). Bäuerlein et al. have recently provided evidence for sufficient vitrification of large volumes samples using a plunge freezer. Drosophila melanogaster larvae nervous systems were excised, incubated in 10% glycerol, and then plunge frozen onto a support grid. Samples were then directly FIB-milled and then imaged. Their results provide evidence of proper vitrification throughout the sample relative to PBS-only controls. This approach opens up the possibility of directly plunge freezing large-volume samples onto grids. Further research and applications of preincubation methods will indeed be exciting for this area of method development. The studies discussed here describe notable novel methods and instrumentation suitable for larger multicellular cryo-EM sample preparation.
6 Future applications of cryo-EM methods to large-volume samples
Studying symbiotic systems offers insight into how host and microbe affect overall health of each other, before, during, and after association. Combining tools across different disciplines allows a deep dive into translating the conversation between the host and microbes. Multi-organism symbiotic relationships are complicated to tease apart the interaction between the host and potentially 10 s or 100 s of different species of bacteria. One symbiotic system that overcomes this challenge is the Squid-Vibrio system, a binary association between the Hawaiian bobtail squid, Euprymna scolopes, and the bioluminescent gram-negative marine bacterium, Vibrio fischeri. Upon hatching, the light organ of the bobtail squid is colonized by a single species of bacteria, V. fischeri; however, it remains aposymbiotic in the absence of the appropriate symbiont, offering great experimental control of different symbiotic states. The last 30 + years of research in this system has shown in great detail the timing and location of key symbiotic events throughout the lifetime of the animal, reviewed in detail from the host and microbe perspectives here, respectively (Nyholm & McFall-Ngai 2021; Visick et al. 2021). Multiscale metabolomic (Koch et al. 2020), transcriptomic (Kremer et al. 2013; Moriano-Gutierrez et al. 2019), computational (Nawroth et al. 2017), and light microscopy approaches (Essock-Burns et al. 2020) have brought to light the journey V. fischeri goes through before, during, and after colonization into the light organ of this bobtail squid and the impact on the long term biology of the host. We propose that the light organ of the bobtail squid is particularly well-suited to applications of these novel, advanced techniques in cryo-EM large-volume sample processing. Applications of cryo-EM workflows will provide a near-native, high-resolution lens into the interface of V. fischeri with host tissue through the onset and maintenance of symbiosis. The fundamental simplicity of the Squid-Vibrio relationship makes this system a logical next choice for application of the emerging tools originating from the cryo-EM community, outlined above.
Successful adaptation of cryo-EM workflows to large-volumes samples with simplified symbiotic relationships will lead to application of cryo-EM technology to more complex symbiotic associations. The zebrafish, Danio rerio, model system would be a great candidate for these methods (Burns & Guillemin 2017). The zebrafish system is a powerful model to study host-microbe interactions that offers a myriad of genetic-manipulation tools for the host and its microbial partners, which are both easily maintained in the lab (Douglas 2019). One major challenge with complex symbiotic samples is the identification of potentially many different microbial species within a single sample, especially during FIB-milling. One solution would require the inoculation of the host with differentially fluoro-labelled bacteria and using the correlative imaging technologies outlined above, most notably the setups that allow coincident FIB-milling and light microscopy, e.g. the PIE-scope (Gorelick et al. 2019). A wide range of symbiotic systems would benefit from the capabilities of cryo-EM including, but not limited to: the vinegar fly Drosophila melanogaster (Douglas 2018); the honey bee, Apis mellifera (Raymann & Moran 2018); the freshwater hydroid, Hydra vulgaris (Deines et al. 2017); and also the rapid expansion of available organoid culture systems (Rauth et al. 2021).
7 Conclusions
Cryo-EM is a relatively young field with a large driving force for innovation and technology. The discipline was originally limited in application to samples such as isolated proteins, viruses or small prokaryotic cell. However, it has quickly transitioned to the exciting next frontier for whole animal, multi-organism complex samples. The drive for advancement in this field stems from the development of commercially available machines like HPF, FIB-SEM, correlative microscopes and continued innovation lies with the creative solutions to increase feasibility, throughput, and accessibility.
For the last decade, it is clear that great strides have been made for developing techniques, manufacturing and combining instruments, and compiling workflows to bring artefact-free, native-state, highest resolution imaging capabilities to the metaorganism research arena. Rapid development at every step of the large-volume sample pipeline continuously pushes the boundaries of sample size limitation, providing new opportunities to study unique model systems, and hard-to-reach tissues. While the future is promising, a few challenges remain difficult to overcome. Cryo-EM’s first application in 1998 to our fundamental understanding prokaryotic biology has come a long way. And the applications of these techniques and technologies to the metaorganism and symbioses systems is the next frontier.
References
Akahoshi DT, Bevins CL (2022) Flagella at the Host-Microbe Interface: Key Functions Intersect With Redundant Responses. Front Immunol 13(March):3–7. https://doi.org/10.3389/fimmu.2022.828758
Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlén LPO, Richter K, Blanc NS, Studer D, Dubochet J (2004) Cryo-electron microscopy of vitreous sections. EMBO J 23(18):3583–3588. https://doi.org/10.1038/sj.emboj.7600366
Al-Amoudi A, Studer D, Dubochet J (2005) Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J Struct Biol 150(1):109–121. https://doi.org/10.1016/j.jsb.2005.01.003
Aulner N, Danckaert A, Fernandes J, Nicola MA, Roux P, Salles A, Tinevez JY, Shorte SL (2018) Fluorescence imaging host pathogen interactions: fifteen years benefit of hindsight…. Curr Opin Microbiol 43:193–198. https://doi.org/10.1016/j.mib.2018.03.001
Ayache J, Beaunier L, Boumendil J, Ehret G, Laub D (2019) Sample preparation for transmission electron microscopy. Methods Mol Biol 1897:417–424. https://doi.org/10.1007/978-1-4939-8935-5_33
Bäuerlein FJB, Pastor-Pareja JC, Fernández-Busnadiego R (2021) Cryo-electron tomography of native Drosophila tissues vitrified by plunge freezing. BioRxiv. https://doi.org/10.1101/2021.04.14.437159
Bouchet-Marquis C, Hoenger A (2011) Cryo-electron tomography on vitrified sections: A critical analysis of benefits and limitations for structural cell biology. Micron 42(2):152–162. https://doi.org/10.1016/j.micron.2010.07.003
Burns AR, Guillemin K (2017) The scales of the zebrafish: host–microbiota interactions from proteins to populations. Curr Opin Microbiol 38:137–141. https://doi.org/10.1016/j.mib.2017.05.011
Cao Y, Miller SS, Dornbusch MR, Castle SS, Lenz P, Ferguson J, Sadowsky MJ, Nelson MS, Klatt C, Samac DA (2018) Widespread occurrence of Sinorhizobium meliloti strains with a type IV secretion system. Symbiosis 75(2):81–91. https://doi.org/10.1007/s13199-018-0547-2
Collingro A, Köstlbacher S, Horn M (2020) Chlamydiae in the Environment. Trends Microbiol 28(11):877–888. https://doi.org/10.1016/j.tim.2020.05.020
Deines P, Lachnit T, Bosch TCG (2017) Competing forces maintain the Hydra metaorganism. Immunol Rev 279(1):123–136. https://doi.org/10.1111/imr.12564
Dobro MJ, Melanson LA, Jensen GJ, McDowall AW (2010) Plunge freezing for electron cryomicroscopy. In Methods in Enzymology (1st ed., Vol. 481, Issue C). Elsevier Inc. https://doi.org/10.1016/S0076-6879(10)81003-1
Douglas AE (2018) The Drosophila model for microbiome research Angela. Lab Anim 47(6):157–164. https://doi.org/10.1038/s41684-018-0065-0.The
Douglas AE (2019) Simple animal models for microbiome research. Nat Rev Microbiol 17(12):764–775. https://doi.org/10.1038/s41579-019-0242-1
Essock-Burns T, Bongrand C, Goldman WE, Ruby EG, McFall-Ngai MJ, Graf J (2020) Interactions of symbiotic partners drive the development of a complex biogeography in the squid-vibrio symbiosis. Mbio 11(3):1–18. https://doi.org/10.1128/mBio.00853-20
Fronk DC, Sachs JL (2022) Symbiotic organs: the nexus of host–microbe evolution. Trends Ecol Evol, 1–12. https://doi.org/10.1016/j.tree.2022.02.014
Gilbert SF, Sapp J, Tauber AI (2012) A symbiotic view of life: We have never been individuals. Q R Biol 87(4):325–341. https://doi.org/10.1086/668166
Gorelick S, Buckley G, Gervinskas G, Johnson TK, Handley A, Caggiano MP, Whisstock JC, Pocock R, de Marco A (2019) PIE-scope, integrated cryo-correlative light and FIB/SEM microscopy. Elife 8:1–15. https://doi.org/10.7554/eLife.45919
Harapin J, Börmel M, Sapra KT, Brunner D, Kaech A, Medalia O (2015) Structural analysis of multicellular organisms with cryo-electron tomography. Nat Methods 12(7):634–636. https://doi.org/10.1038/nmeth.3401
Hayles MF, de Winter DAM (2021) An introduction to cryo-FIB-SEM cross-sectioning of frozen, hydrated Life Science samples. J Microsc 281(2):138–156. https://doi.org/10.1111/jmi.12951
Jani AJ, Cotter PA (2010) Type VI Secretion: Not just for pathogenesis anymore. Cell Host Microbe 8(1):2–6. https://doi.org/10.1016/j.chom.2010.06.012
Kelley K, Raczkowski AM, Klykov O, Jaroenlak P, Bobe D, Kopylov M, Eng ET, Bhabha G, Potter CS, Carragher B, Noble AJ (2022) Waffle Method: A general and flexible approach for improving throughput in FIB-milling. Nat Commun 13(1):1–13. https://doi.org/10.1038/s41467-022-29501-3
Koch EJ, Moriano-Gutierrez S, Ruby EG, McFall-Ngai M, Liebeke M (2020) The impact of persistent colonization by Vibrio fischeri on the metabolome of the host squid Euprymna scolopes. J Exp Biol 223(16). https://doi.org/10.1242/jeb.212860
Kremer N, Philipp EER, Carpentier MC, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EAC, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ (2013) Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14(2):183–194. https://doi.org/10.1016/j.chom.2013.07.006
Liedtke J, Depelteau JS, Briegel A (2022) How advances in cryo-electron tomography have contributed to our current view of bacterial cell biology. J Struct Biol X 6:100065. https://doi.org/10.1016/j.yjsbx.2022.100065
Lin J, Xu L, Yang J, Wang Z, Shen X (2021) Beyond dueling : roles of the type VI secretion system in microbiome modulation, pathogenesis and stress resistance. Stress Biol 1–12. https://doi.org/10.1007/s44154-021-00008-z
López-Jiménez AT, Mostowy S (2021) Emerging technologies and infection models in cellular microbiology. Nat Commun 12(1):1–13. https://doi.org/10.1038/s41467-021-26641-w
Mahamid J, Schampers R, Persoon H, Hyman AA, Baumeister W, Plitzko JM (2015) A focused ion beam milling and lift-out approach for site-specific preparation of frozen-hydrated lamellas from multicellular organisms. J Struct Biol 192(2):262–269. https://doi.org/10.1016/j.jsb.2015.07.012
Mcdonald K, Morphew M, Verkade P, Müller-Reichert T (2007) Recent advances in high-pressure freezing : Equipment- and specimen- loading methods (Issue February 2015). https://doi.org/10.1007/978-1-59745-294-6
McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, … Wernegreen JJ (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA, 110(9), 3229–3236. https://doi.org/10.1073/pnas.1218525110
Milne JLS, Subramaniam S (2009) Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat Rev Microbiol 7(9):666–675. https://doi.org/10.1038/nrmicro2183
Moor H (1987) Theory and Practice of High Pressure Freezing. Cryotechniques Biol Electron Microsc 175–191. https://doi.org/10.1007/978-3-642-72815-0_8
Moriano-Gutierrez S, Koch EJ, Bussan H, Romano K, Belcaid M, Rey FE, Ruby EG, McFall-Ngai MJ (2019) Critical symbiont signals drive both local and systemic changes in diel and developmental host gene expression. Proc Natl Acad Sci USA 116(16):7990–7999. https://doi.org/10.1073/pnas.1819897116
Nans A, Saibil HR, Hayward RD (2014) Pathogen-host reorganization during Chlamydia invasion revealed by cryo-electron tomography. Cell Microbiol 16(10):1457–1472. https://doi.org/10.1111/cmi.12310
Nawroth JC, Guo H, Koch E, Heath-Heckman EAC, Hermanson JC, Ruby EG, Dabiri JO, Kanso E, McFall-Ngai M (2017) Motile cilia create fluid-mechanical microhabitats for the active recruitment of the host microbiome. Proc Natl Acad Sci 114(36):201706926. https://doi.org/10.1073/pnas.1706926114
Nyholm SV, McFall-Ngai MJ (2021) A lasting symbiosis: how the Hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner. Nat Rev Microbiol 19(10):666–679. https://doi.org/10.1038/s41579-021-00567-y
Oikonomou CM, Jensen GJ (2017) Cellular electron cryotomography: Toward structural biology in situ. Annu Rev Biochem 86:873–896. https://doi.org/10.1146/annurev-biochem-061516-044741
Park D, Lara-Tejero M, Waxham MN, Li W, Hu B, Galán JE, Liu J (2018) Visualization of the type III secretion mediated salmonella–host cell interface using cryo-electron tomography. Elife 7:1–15. https://doi.org/10.7554/eLife.39514
Parmenter CD, Nizamudeen ZA (2021) Cryo-FIB-lift-out: practically impossible to practical reality. J Microsc 281(2):157–174. https://doi.org/10.1111/jmi.12953
Pilhofer M, Aistleitner K, Ladinsky MS, König L, Horn M, Jensen GJ (2014) Architecture and host interface of environmental chlamydiae revealed by electron cryotomography. Environ Microbiol 16(2):417–429. https://doi.org/10.1111/1462-2920.12299
Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ (2010) Bacterial TEM. New insights from cryo-microscopy. In Methods in Cell Biology (Vol. 96, Issue C). Elsevier Inc. https://doi.org/10.1016/S0091-679X(10)96002-0
Rauth S, Karmakar S, Batra SK, Ponnusamy MP (2021) Recent Advances in Organoid Development and Applications in Disease Modeling. In Biochimica et Biophysica Acta - Reviews on Cancer (Vol. 1875, Issue 2). https://doi.org/10.1016/j.bbcan.2021.188527.Recent
Raymann K, Moran NA (2018) The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci 26:97–104. https://doi.org/10.1016/j.cois.2018.02.012.The
Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD (2014) A type VI secretion-related pathway in bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16(2):227–236. https://doi.org/10.1016/j.chom.2014.07.007
Sharma V, von Ossowski I, Krishnan V (2021) Exploiting pilus-mediated bacteria-host interactions for health benefits. Mol Aspects Med 81(July):100998. https://doi.org/10.1016/j.mam.2021.100998
Studer D, Graber W, Al-Amoudi A, Eggli P (2001) A new approach for cryofixation by high-pressure freezing. J Microsc 203(3):285–294. https://doi.org/10.1046/j.1365-2818.2001.00919.x
Studer D, Humbel BM, Chiquet M (2008) Electron microscopy of high pressure frozen samples: Bridging the gap between cellular ultrastructure and atomic resolution. Histochem Cell Biol 130(5):877–889. https://doi.org/10.1007/s00418-008-0500-1
Turk M, Baumeister W (2020) The promise and the challenges of cryo-electron tomography. FEBS Lett 594(20):3243–3261. https://doi.org/10.1002/1873-3468.13948
Visick KL, Stabb EV, Ruby EG (2021) A lasting symbiosis: how Vibrio fischeri finds a squid partner and persists within its natural host. Nat Rev Microbiol 19(10):654–665. https://doi.org/10.1038/s41579-021-00557-0
Acknowledgements
This work was supported by funding from the Gordon and Betty Moore Foundation (Symbiosis Model Systems, Grant #9328).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gundlach, K.A., Briegel, A. Zooming in on host-symbiont interactions: advances in cryo-EM sample processing methods and future application to symbiotic tissues. Symbiosis 87, 67–75 (2022). https://doi.org/10.1007/s13199-022-00859-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-022-00859-8