Abstract
Work is in progress for the completion of a computer-aided key to all lichens known to occur in Italy, which will be freely available online, and as a free application for mobile devices. A first example, concerning the lichens of Northern Italy (2.339 infrageneric taxa), is already available online for testing. A computer-generated but manually edited dichotomous key is invoked for all species previously filtered via a multi-entry interface, where several selected characters can be specified in a single step. To optimize the two query interfaces, two different datasets are used, one for the dichotomous, the other for the multi-entry interface.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Traditionally, most identification tools for lichens were paper-published as dichotomous, or more rarely polytomous keys. The structure of such keys typically consists of a series of alternative statements, called “leads”. All leads that need to be evaluated for a single decision form a “couplet”. Dichotomous keys are a special case of polytomous keys, and a key may be also a mixture of simple polytomous and complex dichotomous choices. The generalizing term “single-access key” is used here to include both dichotomous and polytomous keys. The equivalent term in computer science is “decision tree” (Hagedorn et al. 2010).
The advent of computers has allowed the generation of other types of keys, such as free-access and multi-entry keys (Hagedorn et al. 2010). Free-access keys (also known as matrix keys) are alternative to single-access keys. Whereas in a single-access key the sequence of choices (decisions) is fixed by the author (providing a single path to each result), in a free-access key it is up to users, which, at every step, can select from a list one character state at a time. Thus, a free-access key is the set of all possible single-access keys that arise by permutating the order of characters. Although printable free-access keys do exist, they are most suitable for computer-aided identification tools, and have a long development history. Examples are DELTA-IntKey (Dallwitz et al. 2000 onwards), NaviKey (Neubacher and Rambold 2005 onwards), and Xper3 (Vignes Lebbe et al. 2016). Well-known free-access key for lichens are those provided by LIAS (Rambold et al. 2014).
While in a free-access key users must select a single character state at each step of the identification process, multi-entry keys permit to use several characters at the same time, via a multi-character query-form. This first step can be followed by either a field-guide-like page, illustrating the remaining taxa, or (as in the case of our keys) by a dynamically generated single-access key to those taxa only.
After the completion of a new checklist of the lichens of Italy (Nimis 2016), and its incorporation into ITALIC, the online information system on the lichens of Italy (Nimis and Martellos 2002; Martellos 2012), work has started on the preparation of a computer-aided lichen flora of the country, containing keys and descriptions. A first comprehensive key, including all species known from Northern Italy (2.239 infrageneric taxa) has been already published online for testing (Nimis 2019). Separate, nationwide dichotomous keys to given families and/or genera are also being produced, which will be eventually integrated into a complete national key; they are available online at: http://italic.units.it/?procedure=idkeys. The present paper details the main features of the new keys, which are produced using FRIDA, an original software (Martellos 2010) that generates both multi-entry and classical dichotomous keys, integrated into a single query system.
2 The choice of the query interfaces
The creation of a computer-aided identification tool for all lichens of Italy is a challenging task, involving an understanding of principles, technologies, domain knowledge and terminology, as well as pedagogical skills. We have addressed several issues related to the usability of online biodiversity resources by analysing feedback from users, customising the query systems and interfaces accordingly. Feedback was collected from a wide range of users, differing in age, skills, background and interests, in the framework of the European Projects KeyToNature (Nimis 2010), VIBRANT (Dave and Smith 2010), Open Discovery Space (http://opendiscoveryspace.eu) and SiiT (http://www.siit.eu). The main outcome, mainly based on Focus Groups involving 676 persons and on more than 25.000 questionnaires, brought to a new design of the query interfaces. The main results of the testing activity, as summarized by Martellos and Nimis (2015), were:
-
1)
The dichotomous interface was perceived by users as the most user-friendly, and the most effective in identifying closely related taxa, especially in critical groups where species are often distinguished by a combination of characters (i.e. by complex Boolean statements involving multiple characters), rather than by a single character. Its main drawback is the impossibility to proceed in the identification pathway when a specimen lacks one or more characters whose observation is required.
-
2)
The free-access interface was perceived as the most unfriendly, users being often confused by the high number of available characters to choose from, and annoyed by the fact that after each choice they had to re-start the process with a new character; it was also found rather poor for distinguishing among closely related taxa, since it uses a single character at a time; however, the opportunity of having more freedom in the choice of characters was seen as a potential advantage.
-
3)
The multi-entry interface was appreciated for greatly reducing the number of remaining taxa in a single step, although by itself it does not always permit to achieve an identification at species level.
We tried to transfer the input from users into a new and effective product, by combining the capacity of a multi-entry interface of reducing the list of taxa in a single step, with the higher performance of dichotomous keys in dealing with closely related taxa. In our system, a computer-generated but manually edited dichotomous key is invoked for the species which have been previously filtered via a multi-entry interface, which greatly reduces the number of passages required for identification. The graphic outlines of both the multi-entry and the dichotomous interfaces were also designed and tested on the basis of feedback from users.
3 The software
FRIDA (FRiendly IDentificAtion, see Martellos 2010) is a software package for producing digital identification keys, developed since 2003 at the Department of Life Sciences of the University of Trieste, in the framework of project Dryades (Nimis et al. 2003). The software allows users to store and organise characters and their states, taxa, digital images, and textual notes. These resources are used for generating digital identification keys, which can be edited, refined, enriched by further content, and published online, or used through an app for mobile devices (Nimis and Martellos 2009). To allow collaborative efforts in the development of identification keys, FRIDA is based on a double-level architecture, which permits several authors to contribute to a common project, while maintaining a high degree of independence (Martellos 2010). Contrary to most available software for the creation of identification tools, FRIDA was mainly focused on the optimization of classical dichotomous keys. Multi-entry query interfaces were added later, as a consequence of feedback from users. The code of the FRIDA software is available for free upon request to the authors. It comes together with an empty MySQL database with all necessary data tables. Anybody can have it, and modify it as they prefer. A printable reference manual does not exist yet, but but scripts are commented in detail.
4 The data
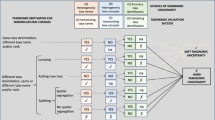
The keys are produced starting from a series of interoperable databases. Figure 1 shows an entity-relationship diagram illustrating the relationships among the main databases involved in the generation, storage, and use of the digital identification keys.
The nomenclatural backbone is provided by the nomenclatural database of ITALIC, which also includes a Thesaurus of several thousands synonyms. Each record in the other databases can be related to 0 or 1 taxon names in ITALIC, since these can contain taxa which do not occur in Italy, being thus absent from ITALIC. Each taxon name in ITALIC can be related to 0 to many records in each database, except the one used for the multi-entry interface, for which the relationship is 0 or 1, each taxon being described by a single record only (see later). Nomenclatural changes which are operated on ITALIC automatically occur in all other databases and in all keys generated by the system.
Due to the high variability of several characters in lichens, a single dataset is not appropriate for serving as a base for generating both a dichotomous key and a multi-entry query interface. For example, a chemically variable species may produce very different spot-tests; if other, less variable characters are available to distinguish that species from all others, it would have little sense to create, in a dichotomous key, as many entries as the number of chemical strains. The best solution would be to construct a key based on a dataset in which spot-tests are not specified for that species, and hence not taken into consideration in the dichotomous pathway. On the contrary, all states of all characters selected for the multi-entry interface should be specified, i.e. all possible spot-tests of a chemically variable species should be databased. For this reason, our system uses two separate datasets, one for the generation of dichotomous keys, the other for creating a multi-entry query interface. In the latter, several character states can be specified for each taxon, e.g. thalli of the same species can react K-, K+ yellow, K+ red, etc.
In the current version of FRIDA (Martellos 2010), data are stored in a MySQL database, and organized into two datasets:
-
1)
In the first dataset, used for producing dichotomous keys, given a set of characters, the infra-taxonomic diversity of each taxon is described by 1 to n records. Each record is an array of couplets character:state (e.g. thallus:crustose). In a record, each character can have a single state only. Records of the same taxon differ for the state of 1 to m characters, m being the total number of characters. These records are used for the production of dichotomous keys. When generating a key, the records are used independently, as if they were different objects. At the end of the generation process, a taxon appears n times in the decision tree. The position of each record in the resulting decision tree is due to its specific array of couplets character:state. High redundancy is allowed in this dataset, since the records of a taxon normally differ for one or a few character states only.
-
2)
In the second dataset, used for producing multi-entry query interfaces, the infra-taxonomic variability of each taxon is described by an array of characters. For each character, from 0 (null) to x states are given, with x the maximum number of states of the character. In this case, a taxon is described by a multidimensional array, in which for each character there is an array of states. Since this dataset is not used for generating decision trees, but for running the multi-entry query system (see below), all the possible infra-taxonomic diversity can be described in a single record.
The relationship between records in the multi-entry database and the one used for the generation of dichotomous keys is 1 to 1 to many, because for each record in the multi-entry database there can be 1 or more records in the key-generating database.
The decision of developing and maintaining two different datasets for the same taxa in the FRIDA system is due to the different purposes they serve. The first one is used for optimizing the decision trees (dichotomous keys), the second for creating a performing multi-entry query interface.
Further data are the descriptions of species, which also derive from ITALIC. We have preferred to avoid computer-generated descriptions, whose shortcomings are well-known. All descriptions are textual, and based on different sources, including original descriptions, recent monographs, and the direct observation of herbarium material, when necessary. The characters typical of a genus are repeated for every single species of that genus. The reason is that the general key to the lichens of Italy will not follow a classical, paper-printed taxonomic scheme where the characters of genera can be specified in a separate paragraph. Furthermore, from that key it will be possible to rapidly derive a high number of smaller keys (e.g. to the lichens of a natural park, of a region, of a school garden etc.) where species of different genera will key out in the same couplet.
Images of species derive from the image database of ITALIC curated by P.L. Nimis & F. Schumm, which presently includes more than 35.000 digital images for almost 5.500 infrageneric taxa, including many species not occurring in Italy. The relationship between the database in which digital keys are stored after generation and refinement and the image archive is 0 to many (see Fig. 1), as the image archive can contain images of taxa which are not included in any identification key, while some taxa can lack images.
Distribution maps, also deriving from ITALIC, will be included into the keys in the next future.
5 The first example
In order to test the query systems, a first comprehensive key has been already published online, which includes all lichens known from Northern Italy, for a total of 2.339 infrageneric taxa (Nimis 2019). Users can access the key by means of two query interfaces:
-
1
The simplest one is a single entry query interface, which follows the dichotomous key from the beginning to the end. This interface allows to generate a list of taxa, or a printable key to the remaining taxa, at every step of the identification process. The usability of the dichotomous keys is greatly enhanced by the availability of illustrations for characters states, and species.
-
2
The second interface, which is shown by default, allows more complex queries. It displays a multi-entry interface (Fig. 2) with a series of selected illustrated characters (e.g. substrates, growth-forms, vegetative propagules, ascocarp-type, photobionts, types of spores, numbers of spores per ascus, etc.), each with 2 to n states. Users can choose, at the same time, one character state for several characters. The query activates a set of algorithms which reduce the original dichotomous key to a smaller one, containing only the taxa matching the query parameters. The original dichotomous key is disrupted and rebuilt, with less steps, but retaining the original sequence of steps chosen by the author(s). The output is a single-entry interface which now works on the new, reduced list of species. If the genus is already known, one can also obtain a dichotomous key of all species of that genus, or can combine the genus name with some other character: e.g. a key to all epilithic species of Calogaya with a placodioid thallus (Fig. 3).
Once the identification process is completed, the system leads to a taxon page which displays the name of the taxon, a textual description, an ecological-distributional note (at the moment present only in the dichotomous keys to genera), and all available pictures with their metadata. All of these items are automatically retrieved from the archives of ITALIC.
The system also permits to produce printable illustrated dichotomous keys “on demand” (Fig. 3) at any step of the identification process (Nascimbene et al. 2010). Examples of paper-published keys largely produced by FRIDA are the key to terricolous lichens of Italy (Nimis and Martellos 2004), the key to macrolichens of Estonia (Randlane et al. 2011) and a key to 100 common lichens of Thailand which is being used to organize lichen labs at the university level (Nimis et al. 2017).
Finally, dichotomous, stand-alone versions of the keys are freely downloadable on smartphones and tablets via the free app KeyToNature (Riccamboni et al. 2010; Nimis et al. 2015), available both for Android and iOS devices.
6 Discussion
Computer-aided keys have several advantages when compared to traditional paper-printed keys:
-
1)
They can be corrected and updated in real time, permitting to add/exclude species, fix eventual mistakes, and update nomenclature without the need of a new paper-printed edition.
-
2)
They are available, with different query interfaces, on different media (online, stand-alone, for mobile devices, paper-printable).
-
3)
They can make use of a potentially unlimited amount of multimedia resources, such as digital images of species and characters, distribution maps, etc. (but for e.g. animals also videos, sound recordings, etc.).
-
4)
They permit the rapid production of smaller keys, devoted to the organisms of a given area or a given habitat (e.g. keys at the regional level, keys to natural parks, keys to species used in biomonitoring studies, simplified keys for school projects, etc.), which can be easily customized to the needs of different potential users.
-
5)
They have a much wider potential outreach than paper-printed books.
The key to the lichens of Northern Italy, which already includes c. 84% of the infrageneric taxa known to occur in the country, is presently in the phase of advanced testing, being continuously corrected and updated online. Further work will be focused in adding other characters to the multi-entry query interface. Nationwide keys to different genera and groups of genera, produced in collaboration with several specialists, are presently being progressively published online in the taxon pages of ITALIC devoted to genera. Once these will be completed, they will be integrated into a general key to all lichens of Italy, whose publication online and in paper-form is foreseen by 2025.
References
Dallwitz MJ, Paine TA, Zurcher EJ (2000 onwards). Principles of interactive keys. http://deltaintkey.com. Accessed 26 June 2020
Dave R, Smith V (2010) ViBRANT—virtual biodiversity research and access network for taxonomy. In: Nimis PL, Vignes Lebbe R (eds): tools for identifying biodiversity: Progress and problems., EUT, Trieste, p. 53
Hagedorn G, Rambold G, Martellos S. (2010) Types of identification keys. In: Nimis PL and Vignes–Lebbe R (eds.), Tools for identifying biodiversity: Progress and problems, EUT Trieste, pp. 59–64
Martellos S (2010) Multi-authored interactive identification keys: the FRIDA (friendly IDentificAtion) package. Taxon 59(3):922–929
Martellos S (2012) From a textual checklist to an information system: the case study of ITALIC, the information system on Italian lichens. Plant Biosystems 146(4):764–770
Martellos S, Nimis PL (2015) From local checklists to online identification portals: a case study on vascular plants. Plos one 10(3):1–18
Nascimbene J, Martellos S, Nimis PL (2010) An integrated system for producing user-specific keys on demand: an application to Italian lichens. In: Nimis PL, Vignes Lebbe R (eds): tools for identifying biodiversity: Progress and problems., EUT, Trieste, pp. 151-156
Neubacher D, Rambold G (2005 onwards). NaviKey – a Java applet and application for accessing descriptive data coded in DELTA format: http://www.navikey.net. Accessed 26 June 2020
Nimis PL (2010) A key to nature. International Innovation 2020:20–22
Nimis PL (2016) The lichens of Italy. A Second Annotated Catalogue. EUT, Trieste, ISBN 9789992037542, 739 pp.
Nimis PL (2019) The lichens of northern Italy. An interactive guide. Continuously updated online at: http://italic.units.it/flora/index.php?procedure=ext_key_home&key_id=241 Accessed 30 June 2020
Nimis PL, Martellos S (2002) Italic, the information system on Italian lichens. In: Llimona X, Lumbsch HT, Ott, S (eds.): Progress and Problems in Lichenology at the Turn of the Millenium. Bibliotheca Lichenologica, J. Cramer, Berlin, Stuttgart, pp. 271-823
Nimis PL, Martellos S (2004) Keys to the lichens of Italy. I. Terricolous species. Ed. Goliardiche, Trieste, ISBN 9788888171739, 341 pp.
Nimis PL, Martellos S (2009) Computer-aided tools for identifying organisms and their importance for protected areas. Journal of Eco Mont 1(2):55–60. https://doi.org/10.1553/ecomont2s61
Nimis PL, Martellos S, Moro A (2003) Il progetto Dryades: come identificare una piante, da Gutenberg a internet. Biologi Italiani 7:9–15
Nimis PL, Riccamboni R, Martellos S (2015) Identification keys on mobile devices: the Dryades experience. Plant Biosystems 146(4):783–788
Nimis PL, Aptroot A, Boonpragob K, Buaruang K, Poengsungnoen V, Polyiam W, Vongshewarat K, Meesim S, Boonpeng C, Phokaeo S, Molsil M, Nirongbutr P, Sangvichien E, Moro A, Pittao E, Martellos S (2017) 100 lichens from Thailand: a tutorial for students. EUT Trieste, ISBN 9788883038532, 123 pp.
Rambold G, Elix JA, Heindl-Tenhunen B, Köhler T, Nash TH III, Neubacher D, Reichert W, Zedda L, Triebel D (2014) LIAS light. Towards the ten thousand species milestone, Mycokeys 8:11–16
Randlane T, Saag A, Martin L, Timdal E, Nimis PL (2011) Epiphytic macrolichens of Estonia. Tartu Ülikooli Kirjastus ISBN: 9789949196524, 326 pp.
Riccamboni R, Mereu A, Boscarol C (2010) Keys to nature: a test on the iPhone market. In: Nimis PL, Vignes Lebbe R (eds): Tools for identifying biodiversity: Progress and problems., EUT, Trieste, pp. 445–450
Vignes Lebbe R, Chesselet P, Diep Thi M-H (2016) Xper3: new tools for collaborating, training and transmitting knowledge on botanical phenotypes. In: Rakotoarisoa NR, Blackmore S, Riera B (eds): botanists of the twenty-first century: roles, challenges and opportunities. UNESCO, Paris. ISBN 9789231001208. pp. 228–239
Acknowledgments
Open access funding provided by Università degli Studi di Trieste within the CRUI-CARE Agreement. Thanks are due to Dr. E. Pittao and Dr. A. Moro (University of Trieste) for their help in the preparation of the article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nimis, P.L., Martellos, S. Towards a digital key to the lichens of Italy. Symbiosis 82, 149–155 (2020). https://doi.org/10.1007/s13199-020-00714-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-020-00714-8