Abstract
Background
Atherosclerosis is a major contributor to morbidity and mortality worldwide. Although several molecular markers associated with atherosclerosis have been developed in recent years, the lack of robust evidence hinders their clinical applications. For these reasons, identification of novel and robust biomarkers will directly contribute to atherosclerosis management in the context of predictive, preventive, and personalized medicine (PPPM). This integrative analysis aimed to identify critical genetic markers of atherosclerosis and further explore the underlying molecular immune mechanism attributing to the altered biomarkers.
Methods
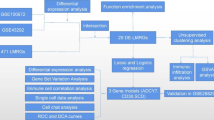
Gene Expression Omnibus (GEO) series datasets were downloaded from GEO. Firstly, differential expression analysis and functional analysis were conducted. Multiple machine-learning strategies were then employed to screen and determine key genetic markers, and receiver operating characteristic (ROC) analysis was used to assess diagnostic value. Subsequently, cell-type identification by estimating relative subsets of RNA transcript (CIBERSORT) and a single-cell RNA sequencing (scRNA-seq) data were performed to explore relationships between signatures and immune cells. Lastly, we validated the biomarkers’ expression in human and mice experiments.
Results
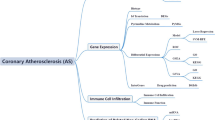
A total of 611 overlapping differentially expressed genes (DEGs) included 361 upregulated and 250 downregulated genes. Based on the enrichment analysis, DEGs were mapped in terms related to immune cell involvements, immune activating process, and inflaming signals. After using multiple machine-learning strategies, dehydrogenase/reductase 9 (DHRS9) and protein tyrosine phosphatase receptor type J (PTPRJ) were identified as critical biomarkers and presented their high diagnostic accuracy for atherosclerosis. From CIBERSORT analysis, both DHRS9 and PTPRJ were significantly related to diverse immune cells, such as macrophages and mast cells. Further scRNA-seq analysis indicated DHRS9 was specifically upregulated in macrophages of atherosclerotic lesions, which was confirmed in atherosclerotic patients and mice.
Conclusions
Our findings are the first to report the involvement of DHRS9 in the atherogenesis, and the proatherogenic effect of DHRS9 is mediated by immune mechanism. In addition, we confirm that DHRS9 is localized in macrophages within atherosclerotic plaques. Therefore, upregulated DHRS9 could be a novel potential target for the future predictive diagnostics, targeted prevention, patient stratification, and personalization of medical services in atherosclerosis.








Similar content being viewed by others
Data availability
The data involved in this study had been described in detail in the “Materials and methods.” The data analyzed during the human and mice experiments are available from the corresponding author on reasonable request.
Code availability
All software applications used are included in this article.
Abbreviations
- ABI :
-
Ankle brachial index
- ASCVD :
-
Atherosclerotic cardiovascular diseases
- AUC :
-
Area under the curve
- BP :
-
Biological process
- CC :
-
Cellular component
- CIBERSORT :
-
Cell-type identification by estimating relative subsets of RNA transcript
- c-IMT :
-
Carotid artery intima medial thickness
- DAPI :
-
4’,6-Diamidino-2-phenylindole
- DEGs :
-
Differentially expressed genes
- DHRS9 :
-
Dehydrogenase/reductase 9
- GAPDH :
-
Anti-glyceraldehyde-3-phosphate dehydrogenase
- GEO :
-
Gene Expression Omnibus
- GO :
-
Gene ontology
- GSE :
-
Gene Expression Omnibus series
- Hcy :
-
Homocysteine
- KEGG :
-
Kyoto encyclopedia of genes and genomes
- LASSO :
-
Least absolute shrinkage and selection operator
- lncRNA :
-
Long non-coding RNA
- MF :
-
Molecular function
- miRNA :
-
MicroRNA
- NCBI :
-
National Center of Biotechnology Information-GEO
- NCD :
-
Normal chow diet
- PCA :
-
Principal component analysis
- PPPM :
-
Predictive, preventive, and personalized medicine
- PTPRJ :
-
Protein tyrosine phosphatase receptor type J
- PWV :
-
Pulse wave velocity
- RF :
-
Random forests
- ROC :
-
Receiver operating characteristic
- scRNA-seq :
-
Single-cell RNA sequencing
- TF :
-
Transcription factor
- UMAP :
-
Uniform Manifold Approximation and Projection
- WD :
-
Western diet
- WGCNA :
-
Weighted gene co-expression network analysis
References
Gallino A, Aboyans V, Diehm C, Cosentino F, Stricker H, Falk E, et al. Non-coronary atherosclerosis. Eur Heart J. 2014;35:1112–9. https://doi.org/10.1093/eurheartj/ehu071.
Ross R. Atherosclerosis–an inflammatory disease. N Engl J Med. 1999;340:115–26. https://doi.org/10.1056/NEJM199901143400207.
Timmis A, Townsend N, Gale CP, Torbica A, Lettino M, Petersen SE, et al. European Society of Cardiology: Cardiovascular Disease Statistics 2019 (Executive Summary). Eur Heart J Qual Care Clin Outcomes. 2020;6:7–9. https://doi.org/10.1093/ehjqcco/qcz065.
Murray CJ, Lopez AD. Measuring the global burden of disease. N Engl J Med. 2013;369:448–57. https://doi.org/10.1056/NEJMra1201534.
Golubnitschaja O, Costigliola V. General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European Association for Predictive. Prev Personal Med EPMA J. 2012;3:14. https://doi.org/10.1186/1878-5085-3-14.
Golubnitschaja O, Watson ID, Topic E, Sandberg S, Ferrari M, Costigliola V. Position paper of the EPMA and EFLM: a global vision of the consolidated promotion of an integrative medical approach to advance health care. EPMA J. 2013;4:12. https://doi.org/10.1186/1878-5085-4-12.
Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, et al. Medicine in the early twenty-first century: paradigm and anticipation - EPMA position paper 2016. EPMA J. 2016;7:23. https://doi.org/10.1186/s13167-016-0072-4.
Ridker PM. A test in context: high-sensitivity c-reactive protein. J Am Coll Cardiol. 2016;67:712–23. https://doi.org/10.1016/j.jacc.2015.11.037.
Kaptoge S, Seshasai SR, Gao P, Freitag DF, Butterworth AS, Borglykke A, et al. Inflammatory cytokines and risk of coronary heart disease: new prospective study and updated meta-analysis. Eur Heart J. 2014;35:578–89. https://doi.org/10.1093/eurheartj/eht367.
Ridker PM. From C-reactive protein to interleukin-6 to interleukin-1: moving upstream to identify novel targets for atheroprotection. Cir Res. 2016;118:145–56. https://doi.org/10.1161/CIRCRESAHA.115.306656.
Koklesova L, Mazurakova A, Samec M, Biringer K, Samuel SM, Büsselberg D, et al. Homocysteine metabolism as the target for predictive medical approach, disease prevention, prognosis, and treatments tailored to the person. EPMA J. 2021;12:1–29. https://doi.org/10.1007/s13167-021-00263-0.
Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016;315:2532–41. https://doi.org/10.1001/jama.2016.5951.
Stitziel NO, Stirrups KE, Masca NG, Erdmann J, Ferrario PG, König IR, et al. Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med. 2016;374:1134–44. https://doi.org/10.1056/NEJMoa1507652.
Döring Y, Noels H, van der Vorst EPC, Neideck C, Egea V, Drechsler M, et al. Vascular CXCR4 limits atherosclerosis by maintaining arterial integrity: evidence from mouse and human studies. Circulation. 2017;136:388–403. https://doi.org/10.1161/CIRCULATIONAHA.117.027646.
Döring Y, van der Vorst EPC, Duchene J, Jansen Y, Gencer S, Bidzhekov K, et al. CXCL12 derived from endothelial cells promotes atherosclerosis to drive coronary artery disease. Circulation. 2019;139:1338–40. https://doi.org/10.1161/CIRCULATIONAHA.118.037953.
Fu Q, Zhao M, Wang D, Hu H, Guo C, Chen W, et al. Coronary plaque characterization assessed by optical coherence tomography and plasma trimethylamine-N-oxide levels in patients with coronary artery disease. Am J Cardiol. 2016;118:1311–5. https://doi.org/10.1016/j.amjcard.2016.07.071.
Lu Y, Zhang X, Hu W, Yang Q. The identification of candidate biomarkers and pathways in atherosclerosis by integrated bioinformatics analysis. Comput Math Methods Med. 2021;2021:6276480. https://doi.org/10.1155/2021/6276480.
Martinez E, Martorell J, Riambau V. Review of serum biomarkers in carotid atherosclerosis. J Vasc Surg. 2020;71:329–41. https://doi.org/10.1016/j.jvs.2019.04.488.
Tibaut M, Caprnda M, Kubatka P, Sinkovič A, Valentova V, Filipova S, et al. Markers of atherosclerosis: part 1 - serological markers. Heart Lung Circ. 2019;28:667–77. https://doi.org/10.1016/j.hlc.2018.06.1057.
Zhang J. Biomarkers of endothelial activation and dysfunction in cardiovascular diseases. Rev Cardiovasc Med. 2022;23:73. https://doi.org/10.31083/j.rcm2302073.
Kobiyama K, Ley K. Atherosclerosis Cir Res. 2018;123:1118–20. https://doi.org/10.1161/CIRCRESAHA.118.313816.
Libby P, Lichtman AH, Hansson GK. Immune effector mechanisms implicated in atherosclerosis: from mice to humans. Immunity. 2013;38:1092–104. https://doi.org/10.1016/j.immuni.2013.06.009.
Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–74. https://doi.org/10.1038/nature01323.
Wolf D, Ley K. Immunity and inflammation in atherosclerosis. Cir Res. 2019;124:315–27. https://doi.org/10.1161/CIRCRESAHA.118.313591.
Soehnlein O, Libby P. Targeting inflammation in atherosclerosis - from experimental insights to the clinic. Nat Rev Drug Discov. 2021;20:589–610. https://doi.org/10.1038/s41573-021-00198-1.
Shen Y, Xu LR, Tang X, Lin CP, Yan D, Xue S, et al. Identification of potential therapeutic targets for atherosclerosis by analysing the gene signature related to different immune cells and immune regulators in atheromatous plaques. BMC Med Genomics. 2021;14:145. https://doi.org/10.1186/s12920-021-00991-2.
Zhao B, Wang D, Liu Y, Zhang X, Wan Z, Wang J, et al. Six-gene signature associated with immune cells in the progression of atherosclerosis discovered by comprehensive bioinformatics analyses. Cardiovasc Ther. 2020;2020:1230513. https://doi.org/10.1155/2020/1230513.
Alsaigh T, Evans D, Frankel D, Torkamani AJb. Decoding the transcriptome of atherosclerotic plaque at single-cell resolution. 2020 BioRxiv. Preprint. https://doi.org/10.1101/2020.03.03.968123
Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, et al. Single-cell immune landscape of human atherosclerotic plaques. Nat Med. 2019;25:1576–88. https://doi.org/10.1038/s41591-019-0590-4.
Depuydt MAC, Prange KHM, Slenders L, Örd T, Elbersen D, Boltjes A, et al. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Cir Res. 2020;127:1437–55. https://doi.org/10.1161/CIRCRESAHA.120.316770.
Steenman M, Espitia O, Maurel B, Guyomarch B, Heymann MF, Pistorius MA, et al. Identification of genomic differences among peripheral arterial beds in atherosclerotic and healthy arteries. Sci Rep. 2018;8:3940. https://doi.org/10.1038/s41598-018-22292-y.
Ayari H, Bricca G. Identification of two genes potentially associated in iron-heme homeostasis in human carotid plaque using microarray analysis. J Biosci. 2013;38:311–5. https://doi.org/10.1007/s12038-013-9310-2.
Döring Y, Manthey HD, Drechsler M, Lievens D, Megens RT, Soehnlein O, et al. Auto-antigenic protein-DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation. 2012;125:1673–83. https://doi.org/10.1161/CIRCULATIONAHA.111.046755.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. https://doi.org/10.1186/gb-2004-5-10-r80.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43: e47. https://doi.org/10.1093/nar/gkv007.
Wickham H. ggplot2: elegant graphics for data analysis. Springer-Verlag New York.
Kolde R. pheatmap: Pretty Heatmaps. R package version 1.0.12. 2019.
Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. https://doi.org/10.1186/1471-2105-12-35.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. https://doi.org/10.1089/omi.2011.0118.
Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–4. https://doi.org/10.1093/bioinformatics/btv300.
Breiman L. Random Forests. Mach Learn. 2001;45:5–32. https://doi.org/10.1023/A:1010933404324.
Tibshirani R. Regression shrinkage and selection via the lasso. J R Statist Soc B. 1996;58:267–88. https://doi.org/10.1111/j.2517-6161.1996.tb02080.x.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. https://doi.org/10.1186/1471-2105-9-559.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. https://doi.org/10.1186/1471-2105-12-77.
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-7. https://doi.org/10.1093/nar/gkt1248.
Zhang Q, Liu W, Zhang HM, Xie GY, Miao YR, Xia M, et al. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets. Genomics Proteomics Bioinforma. 2020;18:120–8. https://doi.org/10.1016/j.gpb.2019.09.006.
Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46:D360-d70. https://doi.org/10.1093/nar/gkx1144.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. https://doi.org/10.1101/gr.1239303.
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20. https://doi.org/10.1038/nbt.4096.
Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–72. https://doi.org/10.1038/s41590-018-0276-y.
Carter HE, Schofield D, Shrestha R. Productivity costs of cardiovascular disease mortality across disease types and socioeconomic groups. Open heart. 2019;6:e000939. https://doi.org/10.1136/openhrt-2018-000939.
Ergin A, Muntner P, Sherwin R, He J. Secular trends in cardiovascular disease mortality, incidence, and case fatality rates in adults in the United States. Am J Med. 2004;117:219–27. https://doi.org/10.1016/j.amjmed.2004.03.017.
Adams A, Bojara W, Schunk K. Early diagnosis and treatment of coronary heart disease in symptomatic subjects with advanced vascular atherosclerosis of the carotid artery (type III and IV b findings using ultrasound). Cardiol Res. 2017;8:7–12. https://doi.org/10.14740/cr516w.
Abdolmaleki F, Gheibi Hayat SM, Bianconi V, Johnston TP, Sahebkar A. Atherosclerosis and immunity: a perspective. Trends Cardiovasc Med. 2019;29:363–71. https://doi.org/10.1016/j.tcm.2018.09.017.
Du M, Wang X, Mao X, Yang L, Huang K, Zhang F, et al. Absence of Interferon regulatory factor 1 protects against atherosclerosis in apolipoprotein E-deficient mice. Theranostics. 2019;9:4688–703. https://doi.org/10.7150/thno.36862.
Sozen E, Karademir B, Yazgan B, Bozaykut P, Ozer NK. Potential role of proteasome on c-jun related signaling in hypercholesterolemia induced atherosclerosis. Redox Biol. 2014;2:732–8. https://doi.org/10.1016/j.redox.2014.02.007.
Everts HB, Silva KA, Montgomery S, Suo L, Menser M, Valet AS, et al. Retinoid metabolism is altered in human and mouse cicatricial alopecia. J Invest Dermatol. 2013;133:325–33. https://doi.org/10.1038/jid.2012.393.
Chazenbalk G, Chen YH, Heneidi S, Lee JM, Pall M, Chen YD, et al. Abnormal expression of genes involved in inflammation, lipid metabolism, and Wnt signaling in the adipose tissue of polycystic ovary syndrome. J Clin Endocrinol Metab. 2012;97:E765–70. https://doi.org/10.1210/jc.2011-2377.
Zhang D, Li Z, Zhang R, Yang X, Zhang D, Li Q, et al. Identification of differentially expressed and methylated genes associated with rheumatoid arthritis based on network. Autoimmunity. 2020;53:303–13. https://doi.org/10.1080/08916934.2020.1786069.
Hu L, Chen HY, Han T, Yang GZ, Feng D, Qi CY, et al. Downregulation of DHRS9 expression in colorectal cancer tissues and its prognostic significance. Tumour Biol. 2016;37:837–45. https://doi.org/10.1007/s13277-015-3880-6.
Li HB, Zhou J, Zhao F, Yu J, Xu L. Prognostic Impact of DHRS9 Overexpression in pancreatic cancer. Cancer Manag Research. 2020;12:5997–6006. https://doi.org/10.2147/CMAR.S251897.
Shimomura H, Sasahira T, Nakashima C, Shimomura-Kurihara M, Kirita T. Downregulation of DHRS9 is associated with poor prognosis in oral squamous cell carcinoma. Pathology. 2018;50:642–7. https://doi.org/10.1016/j.pathol.2018.06.002.
Belyaeva OV, Wirth SE, Boeglin WE, Karki S, Goggans KR, Wendell SG, et al. Dehydrogenase reductase 9 (SDR9C4) and related homologs recognize a broad spectrum of lipid mediator oxylipins as substrates. J Biol Chem. 2022;298:101527. https://doi.org/10.1016/j.jbc.2021.101527.
Riquelme P, Amodio G, Macedo C, Moreau A, Obermajer N, Brochhausen C, et al. DHRS9 is a stable marker of human regulatory macrophages. Transplantation. 2017;101:2731–8. https://doi.org/10.1097/TP.0000000000001814.
Hutchinson JA, Riquelme P, Geissler EK, Fändrich F. Human regulatory macrophages. Methods Mol Biol. 2011;677:181–92. https://doi.org/10.1007/978-1-60761-869-0_13.
Gertow K, Nobili E, Folkersen L, Newman JW, Pedersen TL, Ekstrand J, et al. 12- and 15-lipoxygenases in human carotid atherosclerotic lesions: associations with cerebrovascular symptoms. Atherosclerosis. 2011;215:411–6. https://doi.org/10.1016/j.atherosclerosis.2011.01.015.
Mallat Z, Nakamura T, Ohan J, Lesèche G, Tedgui A, Maclouf J, et al. The relationship of hydroxyeicosatetraenoic acids and F2-isoprostanes to plaque instability in human carotid atherosclerosis. J Clin Invest. 1999;103:421–7. https://doi.org/10.1172/JCI3985.
Burleigh ME, Babaev VR, Oates JA, Harris RC, Gautam S, Riendeau D, et al. Cyclooxygenase-2 promotes early atherosclerotic lesion formation in LDL receptor-deficient mice. Circulation. 2002;105:1816–23. https://doi.org/10.1161/01.cir.0000014927.74465.7f.
Ylä-Herttuala S, Rosenfeld ME, Parthasarathy S, Glass CK, Sigal E, Witztum JL, et al. Colocalization of 15-lipoxygenase mRNA and protein with epitopes of oxidized low density lipoprotein in macrophage-rich areas of atherosclerotic lesions. Proc Natl Acad Sci U S A. 1990;87:6959–63. https://doi.org/10.1073/pnas.87.18.6959.
Lorenz MW, Markus HS, Bots ML, Rosvall M, Sitzer M. Prediction of clinical cardiovascular events with carotid intima-media thickness: a systematic review and meta-analysis. Circulation. 2007;115:459–67. https://doi.org/10.1161/CIRCULATIONAHA.106.628875.
Ato D. Pitfalls in the ankle-brachial index and brachial-ankle pulse wave velocity. Vasc Health Risk Manag. 2018;14:41–62. https://doi.org/10.2147/VHRM.S159437.
Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–52. https://doi.org/10.1016/S0140-6736(04)17018-9.
Bogiatzi C, Gloor G, Allen-Vercoe E, Reid G, Wong RG, Urquhart BL, et al. Metabolic products of the intestinal microbiome and extremes of atherosclerosis. Atherosclerosis. 2018;273:91–7. https://doi.org/10.1016/j.atherosclerosis.2018.04.015.
Bubnov R, Babenko L, Lazarenko L, Kryvtsova M, Shcherbakov O, Zholobak N, et al. Can tailored nanoceria act as a prebiotic? Report on improved lipid profile and gut microbiota in obese mice. EPMA J. 2019;10:317–35. https://doi.org/10.1007/s13167-019-00190-1.
Park D, Lee JH, Han S. Underweight: another risk factor for cardiovascular disease?: A cross-sectional 2013 Behavioral Risk Factor Surveillance System (BRFSS) study of 491,773 individuals in the USA. Medicine. 2017;96:e8769. https://doi.org/10.1097/MD.0000000000008769.
Golubnitschaja O, Liskova A, Koklesova L, Samec M, Biringer K, Büsselberg D, et al. Caution, “normal” BMI: health risks associated with potentially masked individual underweight-EPMA Position Paper 2021. EPMA J. 2021;12:243–64. https://doi.org/10.1007/s13167-021-00251-4.
Acknowledgements
Authors thank National Natural Science Foundation of China (Grant no: NSFC 82170857) for its support for this study.
Funding
This work was supported by the National Natural Science Foundation of China (Grant no: NSFC 82170857).
Author information
Authors and Affiliations
Contributions
JX and HZ were responsible for data acquisition and analysis. YC conducted the human and mice experiments. JX drafted the manuscript. GX designed the study and revised the article. All the authors read and approved the final manuscript for submission.
Corresponding author
Ethics declarations
Ethics approval
The protocol for collecting human tissue samples was approved by Ethics Committee of the General Hospital of Central Theater Command (approval no. [2021]004–02), and all procedures were performed in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Written informed consent was provided by all participants before their inclusion in the study. The animal experimental protocols were permitted by the Animal Ethics Committee of the General Hospital of Central Theater Command (approval no.2021013).
Consent to participate
All individuals were informed about the purposes of the study and have signed their consent for publishing the data.
Consent for publication
After reviewing the manuscript, all authors agreed with its publication in the current form.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1
PPPM strategies of DHRS9 in atherosclerosis. Abbreviations: DHRS9 dehydrogenase/reductase 9; PPPM: predictive, preventive, and personalized medicine (DOCX 18 KB)
Rights and permissions
About this article
Cite this article
Xu, J., Zhou, H., Cheng, Y. et al. Identifying potential signatures for atherosclerosis in the context of predictive, preventive, and personalized medicine using integrative bioinformatics approaches and machine-learning strategies. EPMA Journal 13, 433–449 (2022). https://doi.org/10.1007/s13167-022-00289-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-022-00289-y