Abstract
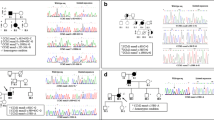
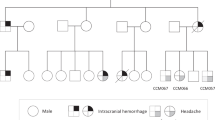
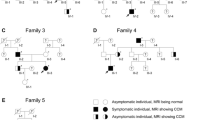
Cerebral cavernous malformation (CCM) is a vascular disease that affects the central nervous system, which familial form is due to autosomal dominant mutations in the genes KRIT1(CCM1), MGC4607(CCM2), and PDCD10(CCM3). Patients affected by the PDCD10 mutations usually have the onset of symptoms at an early age and a more aggressive phenotype. The aim of this study is to investigate the molecular mechanism involved with CCM3 disease pathogenesis. Herein, we report two typical cases of CCM3 phenotype and compare the clinical and neuroradiological findings with five patients with a familial form of KRIT1 or CCM2 mutations and six patients with a sporadic form. In addition, we evaluated the PDCD10 gene expression by qPCR and developed a bioinformatic pipeline to understand the structural changes of mutations. The two CCM3 patients had an early onset of symptoms and a high lesion burden. Furthermore, the sequencing showed that Patient 1 had a frameshift mutation in c.222delT; p.(Asn75Thrfs*14) that leads to lacking the last 124 C-terminal amino acids on its primary structure and Patient 2 had a variant on the splicing site region c.475-2A > G. The mRNA expression was fourfold lower in both patients with PDCD10 mutation. Using in silico analysis, we identify that the frameshift mutation transcript lacks the C-terminal FAT-homology domain compared to the wild-type PDCD10 and preserves the N-terminal dimerization domain. The two patients studied here allow estimating the potential impact of mutations in clinical interpretation as well as support to better understand the mechanism and pathogenesis of CCM3.


Similar content being viewed by others
References
Salman RAS, Hall JM, Horne MA, Moultrie F, Josephson CB, Bhattacharya JJ, et al. Untreated clinical course of cerebral cavernous malformations: A prospective, population-based cohort study. Lancet Neurol Elsevier Ltd. 2012;11:217–24.
Akers A, Al-Shahi Salman R, A Awad I, Dahlem K, Flemming K, Hart B, et al. Synopsis of Guidelines for the Clinical Management of Cerebral Cavernous Malformations: Consensus Recommendations Based on Systematic Literature Review by the Angioma Alliance Scientific Advisory Board Clinical Experts Panel. Neurosurgery. 2017;80:665–80.
De Souza JM, Domingues RC, Cruz LCH, Domingues FS, Iasbeck T, Gasparetto EL. Susceptibility-weighted imaging for the evaluation of patients with familial cerebral cavernous malformations: A comparison with T2-weighted fast spin-echo and gradient-echo sequences. Am J Neuroradiol. 2008;29:154–8.
Labauge P, Denier C, Bergametti F, Tournier-Lasserve E. Genetics of cavernous angiomas. Lancet Neurol [Internet]. Elsevier; 2007;6:237–44. Available from: https://doi.org/10.1016/S1474-4422(07)70053-4
Gault J, Sain S, Hu LJ, Awad IA. Spectrum of genotype and clinical manifestations in cerebral cavernous malformations. Neurosurgery. 2006;59:1278–84.
Denier C, Goutagny S, Labauge P, Krivosic V, Arnoult M, Cousina, et al. Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am J Hum Genet. 2004;74:326–37.
Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76:42–51.
Fontes-Dantas FL, da Fontoura Galvão G, Veloso da Silva E, Alves-Leon S, Cecília da Silva Rêgo C, Garcia DG, et al. Novel CCM1 (KRIT1) Mutation Detection in Brazilian Familial Cerebral Cavernous Malformation: Different Genetic Variants in Inflammation, Oxidative Stress, and Drug Metabolism Genes Affect Disease Aggressiveness. World Neurosurg. United States 2020;535–540.e8. Available from: https://doi.org/10.1016/j.wneu.2020.02.119.
Awad IA, Polster SP. Cavernous angiomas: deconstructing a neurosurgical disease. J Neurosurg. 2019;131:1–13.
D’Angelo R, Marini V, Rinaldi C, Origone P, Dorcaratto A, Avolio M, et al. Mutation analysis of CCM1, CCM2 and CCM3 genes in a cohort of Italian patients with cerebral cavernous malformation. Brain Pathol. 2011;21:215–24.
Taslimi S, Modabbernia A, Amin-Hanjani S, Barker FG, Macdonald RL. Natural history of cavernous malformation. Neurology. 2016;86:1984–91.
Batra S, Lin D, Recinos PF, Zhang J, Rigamonti D. Cavernous malformations: natural history, diagnosis and treatment. Nat Rev Neurol [Internet]. Nature Publishing Group 2009;5:659–70. Available from: http://www.nature.com/doifinder/https://doi.org/10.1038/nrneurol.2009.177
Shenkar R, Shi C, Rebeiz T, Stockton R a, McDonald D a, Mikati AG, et al. Exceptional aggressiveness of cerebral cavernous malformation disease associated with PDCD10 mutations. Hum Vet Med J Bioflux Soc [Internet]. 2014;17:188–96. Available from: http://www.nature.com/gim/journal/v17/n3/full/gim201497a.html#acknowledgments
Bravi L, Rudini N, Cuttano R, Giampietro C, Maddaluno L, Ferrarini L, et al. Sulindac metabolites decrease cerebrovascular malformations in CCM3 -knockout mice. Proc Natl Acad Sci. 2015;112:8421–6.
Zhou HJ, Qin L, Jiang Q, Murray KN, Zhang H, Li B, et al. Caveolae-mediated Tie2 signaling contributes to CCM pathogenesis in a brain endothelial cell-specific Pdcd10-deficient mouse model. Nat Commun. Springer US 2021;12. Available from: https://doi.org/10.1038/s41467-020-20774-0.
Zheng X, Xu C, Di Lorenzo A, Kleaveland B, Zou Z, Seiler C, et al. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J Clin Invest. 2010;120:2795–804.
Voss K, Stahl S, Schleider E, Ullrich S, Nickel J, Mueller TD, et al. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007;8:249–56.
Xu X, Wang X, Zhang Y, Wang DC, Ding J. Structural basis for the unique heterodimeric assembly between cerebral cavernous malformation 3 and germinal center kinase III. Structure Elsevier Ltd. 2013;21:1059–66.
Li X, Zhang R, Zhang H, He Y, Ji W, Min W, et al. Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity. J Biol Chem. 2010;285:24099–107.
Fisher OS, Boggon TJ. Signaling pathways and the cerebral cavernous malformations proteins: Lessons from structural biology. Cell Mol Life Sci. 2014;71:1881–92.
Pombo CM, The GCK. II and III subfamilies of the STE20 group kinases. Front Biosci. 2007;12:850.
Ling P, Lu T-J, Yuan C-J, Lai M-D. Biosignaling of mammalian Ste20-related kinases. Cell Signal. 2008;20:1237–47.
Lu T-J, Lai W-Y, Huang C-YF, Hsieh W-J, Yu J-S, Hsieh Y-J, et al. Inhibition of Cell Migration by Autophosphorylated Mammalian Sterile 20-Like Kinase 3 (MST3) Involves Paxillin and Protein-tyrosine Phosphatase-PEST. J Biol Chem. 2006;281:38405–17.
Preisinger C, Short B, De Corte V, Bruyneel E, Haas A, Kopajtich R, et al. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3ζ. J Cell Biol. 2004;164:1009–20.
Dan I, Ong SE, Watanabe NM, Blagoev B, Nielsen MM, Kajikawa E, et al. Cloning of MASK, a novel member of the mammalian germinal center kinase III subfamily, with apoptosis-inducing properties. J Biol Chem. 2002;277:5929–39.
Huang C-YF, Wu Y-M, Hsu C-Y, Lee W-S, Lai M-D, Lu T-J, et al. Caspase Activation of Mammalian Sterile 20-like Kinase 3 (Mst3). J Biol Chem. 2002;277:34367–74.
Nogueira E, Fidalgo M, Molnar A, Kyriakis J, Force T, Zalvide J, et al. SOK1 translocates from the golgi to the nucleus upon chemical anoxia and induces apoptotic cell death. J Biol Chem. 2008;283:16248–58.
Lant B, Pal S, Chapman EM, Yu B, Witvliet D, Choi S, et al. Interrogating the ccm-3 Gene Network. Cell Rep United States. 2018;24:2857-2868.e4.
Padarti A, Zhang J. Recent advances in cerebral cavernous malformation research. Vessel plus. 2018;2.
Dibble CF, Horst JA, Malone MH, Park K, Temple B, Cheeseman H, et al. Defining the Functional Domain of Programmed Cell Death 10 through Its Interactions with Phosphatidylinositol-3,4,5-Trisphosphate. Hofmann A, editor. PLoS One. 2010;5:e11740.
Oliveira TT, Fontes-Dantas FL, de Medeiros Oliveira RK, Pinheiro DML, Coutinho LG, da Silva VL, et al. Chemical Inhibition of Apurinic-Apyrimidinic Endonuclease 1 Redox and DNA Repair Functions Affects the Inflammatory Response via Different but Overlapping Mechanisms. Front cell Dev Biol. 2021;9:731588. Available from: https://doi.org/10.3389/fcell.2021.731588.
Allhorn S, Böing C, Koch AA, Kimmig R, Gashaw I. TLR3 and TLR4 expression in healthy and diseased human endometrium. Reprod Biol Endocrinol. 2008;6:1–11.
You C, ErolSandalcioglu I, Dammann P, Felbor U, Sure U, Zhu Y. Loss of CCM3 impairs DLL4-Notch signalling: Implication in endothelial angiogenesis and in inherited cerebral cavernous malformations. J Cell Mol Med. 2013;17:407–18.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med United States. 2015;17:405–24.
Tavtigian SV, Harrison SM, Boucher KM, Biesecker LG. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum Mutat. 2020;41:1734–7.
Mcguffin LJ, Adiyaman R, Maghrabi AHA, Shuid AN, Brackenridge DA, Nealon JO, et al. IntFOLD: An integrated web resource for high performance protein structure and function prediction. Nucleic Acids Res Oxford Univ Press. 2019;47:W408–13.
McGuffin LJ, Shuid AN, Kempster R, Maghrabi AHA, Nealon JO, Salehe BR, et al. Accurate template-based modeling in CASP12 using the IntFOLD4-TS, ModFOLD6, and ReFOLD methods. Proteins Struct Funct Bioinforma. 2018;86:335–44.
Wang S, Peng J, Xu J. Alignment of distantly related protein structures: Algorithm, bound and implications to homology modeling. Bioinformatics. 2011;27:2537–45.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:808–15.
Abbasi WA, Yaseen A, Hassan FU, Andleeb S, Minhas FUAA. ISLAND: in-silico proteins binding affinity prediction using sequence information. BioData Min BioData Mining. 2020;13:1–13.
Yan Y, Zhang D, Zhou P, Li B, Huang S-Y. HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45:W365–73.
Liquori CL, Berg MJ, Squitieri F, Ottenbacher M, Sorlie M, Leedom TP, et al. Low frequency of PDCD10 mutations in a panel of CCM3 probands: potential for a fourth CCM locus. Hum Mutat [Internet]. Wiley Subscription Services, Inc., A Wiley Company; 2006;27:118. Available from: https://doi.org/10.1002/humu.9389
Riant F, Cecillon M, Saugier-Veber P, Tournier-Lasserve E. CCM molecular screening in a diagnosis context: novel unclassified variants leading to abnormal splicing and importance of large deletions. Neurogenet United States. 2013;14:133–41.
Cigoli MS, Avemaria F, De Benedetti S, Gesu GP, Accorsi LG, Parmigiani S, et al. PDCD10 gene mutations in multiple cerebral cavernous malformations. PLoS One. 2014;9:e110438.
Merello E, Pavanello M, Consales A, Mascelli S, Raso A, Accogli A, et al. Genetic Screening of Pediatric Cavernous Malformations. J Mol Neurosci [Internet]. J Mol Neurosci 2016;60:232–8. Available from: https://doi.org/10.1007/s12031-016-0806-8
Riant F, Bergametti F, Fournier H-D, Chapon F, Michalak-Provost S, Cecillon M, et al. CCM3 Mutations Are Associated with Early-Onset Cerebral Hemorrhage and Multiple Meningiomas. Mol Syndromol. 2013;4:165–72.
Ramírez-Aportela E, López-Blanco JR, Chacón P. FRODOCK 2.0: fast protein–protein docking server. Bioinformatics. 2016;32:2386–8.
Guclu B, Ozturk AK, Pricola KL, Bilguvar K, Shin D, O’Roak BJ, et al. Mutations in Apoptosis-related Gene, PDCD10, Cause Cerebral Cavernous Malformation 3. Neurosurgery [Internet]. 2005;57. Available from: https://doi.org/10.1227/01.neu.0000180811.56157.e1.
Anna A, Monika G. Splicing mutations in human genetic disorders: examples, detection, and confirmation. J Appl Genet. 2018;59:253–68.
Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13.
Smeby J, Sveen A, Eilertsen IA, Danielsen SA, Hoff AM, Eide PW, et al. Transcriptional and functional consequences of TP53 splice mutations in colorectal cancer. Oncogenesis. Springer US; 2019;8. Available from: https://doi.org/10.1038/s41389-019-0141-3.
Cavé-Riant F, Denier C, Labauge P, Cécillon M, Maciazek J, Joutel A, et al. Spectrum and expression analysis of KRIT1 mutations in 121 consecutive and unrelated patients with Cerebral Cavernous Malformations. Eur J Hum Genet EJHG [Internet]. 2002;10:733–40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12404106
Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–92.
Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104:173–6.
Aksit MA, Bowling AD, Evans TA, Joynt AT, Osorio D, Patel S, et al. Decreased mRNA and protein stability of W1282X limits response to modulator therapy. J Cyst Fibros Off J Eur Cyst Fibros Soc. 2019;18:606–13.
You KT, Li LS, Kim N-G, Kang HJ, Koh KH, Chwae Y-J, et al. Selective translational repression of truncated proteins from frameshift mutation-derived mRNAs in tumors. PLoS Biol United States. 2007;5:e109.
Cardiero G, Musollino G, Prezioso R, Lacerra G. mRNA Analysis of Frameshift Mutations with Stop Codon in the Last Exon: The Case of Hemoglobins Campania [α1 cod95 (-C)] and Sciacca [α1 cod109 (-C)]. Biomed Switzerland 2021;9.
Baranoski JF, Kalani MYS, Przybylowski CJ, Zabramski JM. Cerebral Cavernous Malformations: Review of the Genetic and Protein-Protein Interactions Resulting in Disease Pathogenesis. Front Surg. 2016;3:3–8.
Fauth C, Rostasy K, Rath M, Gizewski E, Lederer AG, Sure U, et al. Highly variable intrafamilial manifestations of a CCM3 mutation ranging from acute childhood cerebral haemorrhage to late-onset meningiomas. Clin Neurol Neurosurg Elsevier BV. 2015;128:41–3.
Nikoubashman O, Wiesmann M, Tournier-Lasserve E, Mankad K, Bourgeois M, Brunelle F, et al. Natural history of cerebral dot-like cavernomas. Clin Radiol Royal Coll Radiol. 2013;68:e453-9.
Draheim KM, Li X, Zhang R, Fisher OS, Villari G, Boggon TJ, et al. CCM2-CCM3 interaction stabilizes their protein expression and permits endothelial network formation. J Cell Biol. 2015;208:987–1001.
Ceccarelli DF, Laister RC, Mulligan VK, Kean MJ, Goudreault M, Scott IC, et al. CCM3/PDCD10 heterodimerizes with germinal center kinase III (GCKIII) proteins using a mechanism analogous to CCM3 homodimerization. J Biol Chem. 2011;286:25056–64.
Harrington LS, Sainson RCA, Williams CK, Taylor JM, Shi W, Li JL, et al. Regulation of multiple angiogenic pathways by Dll4 and Notch in human umbilical vein endothelial cells. Microvasc Res. 2008;75:144–54.
Qin L, Zhang H, Li B, Jiang Q, Lopez F, Min W, et al. CCM3 Loss-Induced Lymphatic Defect Is Mediated by the Augmented VEGFR3-ERK1/2 Signaling. Arterioscler Thromb Vasc Biol. 2021;41:2943–60.
Narasimhan P, Liu J, Song YS, Massengale JL, Chan PH. VEGF Stimulates the ERK 1/2 Signaling Pathway and Apoptosis in Cerebral Endothelial Cells After Ischemic Conditions. Stroke. 2009;40:1467–73.
Tang AT, Sullivan KR, Hong CC, Goddard LM, Mahadevan A, Ren A, et al. Distinct cellular roles for PDCD10 define a gut-brain axis in cerebral cavernous malformation. Sci Transl Med 2019;11. Available from: https://doi.org/10.1126/scitranslmed.aaw3521.
Denier C, Labauge P, Bergametti F, Marchelli F, Riant F, Arnoult M, et al. Genotype–phenotype correlations in cerebral cavernous malformations patients. Ann Neurol [Internet]. Wiley Subscription Services, Inc., A Wiley Company 2006;60:550–6. Available from: https://doi.org/10.1002/ana.20947
Choquet H, Pawlikowska L, Nelson J, McCulloch CE, Akers A, Baca B, et al. Polymorphisms in Inflammatory and Immune Response Genes Associated with Cerebral Cavernous Malformation Type 1 Severity. Cerebrovasc Dis. 2014;38:433–40.
Yu W, Jin H, You Q, Nan D, Huang Y. A novel PDCD10 gene mutation in cerebral cavernous malformations: a case report and review of the literature. J Pain Res. 2019;12:1127–32.
Acknowledgements
The authors thank the Cavernoma Alliance Brazil Research Institute – Aliança Cavernoma Brasil for the logistic assistance.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Brazilian National Council for Scientific and Technological Development (CNPq Number 440779/2016-2), Senator Romario Faria parliamentary amendment (no. 37990007 EIND), financial support of Coordination for the Improvement of Higher Education Personnel (CAPES Number 88887.130752/2016-00), FAPERJ E-26/210.657/2021, E-26/210.273/2018 and E-26/201.040/2021 and Chamada Pública MCTI/FINEP/CT-INFRA-PROINFRA 02/2014 – Equipamentos Multiusuários – Ref. nº 0097/2016.
Author information
Authors and Affiliations
Contributions
Material preparation and analysis were performed by Gustavo da Fontoura Galvão and Fabrícia Lima Fontes-Dantas. Data collection was performed by Gustavo da Fontoura Galvão and Jorge Marcondes de Souza. Sample processing and lab work was performed by Fabrícia Lima Fontes-Dantas, Elielson Veloso da Silva, and Luisa Menezes Trefilio. The first draft of the manuscript was written by Gustavo da Fontoura Galvão and review by Fabrícia Lima Fontes-Dantas and Jorge Marcondes de Souza. Funding acquisition, resources, and supervision were performed by Soniza Vieira Alves-Leon, Jorge Marcondes de Souza, and Fabrícia Lima Fontes-Dantas. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
Due to the sensitive nature of the data collected, qualified researcher on human subjects conducted this study having been approved by the National Council for Ethics in Research (CAAE 69409617.9.0000.5258).
Data Availability
Not applicable.
Code Availability
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
da Fontoura Galvão, G., da Silva, E.V., Trefilio, L.M. et al. Comprehensive CCM3 Mutational Analysis in Two Patients with Syndromic Cerebral Cavernous Malformation. Transl. Stroke Res. 15, 411–421 (2024). https://doi.org/10.1007/s12975-023-01131-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12975-023-01131-x