Abstract
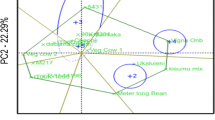
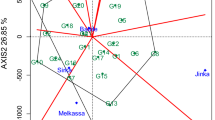
The variable cowpea productivity across different environments demands evaluating the performance of genotypes in a breeding program prior to their release. The aim of this study was to assess yield stability of eight cowpea advanced breeding lines selected from participatory varietal selection in multilocational trials, and to identify mega-environments for cowpea production in Ghana. The genotypes were evaluated across five environments in 2016 and 2017 in randomized complete block design with three replications. The GEA-R version 4.0 software was used for genotype main effect plus genotype by environment interaction (GGE) biplot analyses. Analysis of variance (PROC GLM of SAS using a RANDOM statement with the TEST option) detected significant variations for location, year, genotype, environment, and their interactions. The results showed that the yield performances of the cowpea genotypes were highly influenced by genotype × environment interaction effects. The principal component 1 (PC1) and PC2 were significant components which accounted for 46.75% and 22.84% of GGE sum of squares, respectively. We showed for the first time, two mega-environments for cowpea production and testing in the major cowpea production agro-ecologies in Ghana. The genotypes SARI-6-2-6 and IT07K-303-1 were adapted to Damongo, Nyankpala, and Tumu, whereas SARI-2-50-80 was adapted to Yendi and Manga. The best ranking location was Damongo followed by Tumu, and Nyankpala. The high-yielding genotypes, IT86D-610, IT10K-837-1, IT07K-303-1, and SARI-2-50-80 had significant higher grain yields than the check (Bawutawuta) and were recommended for release as cultivars (or as breeding lines) to boost cowpea production in Ghana.




Similar content being viewed by others
References
Bhartiya A, Aditya JP, Singh K, Purwar JP, Agarwal A (2017) AMMI and GGE biplot analysis of multi environment yield trial of soybean in North Western Himalayan state Uttarakh and of India. Legume Res Int J 40(OF):306–312. https://doi.org/10.18805/lr.v0iOF.3548
Blanche SB, Myers GO (2006) Identifying discriminating locations for cultivar in Louisiana. Crop Sci 46:946–949
Boukar O, Fatokun CA, Huynh BL, Roberts PA, Close TJ (2016) Genomic tools in cowpea breeding programs: status and perspectives. Front Plant Sci 2016(7):1–13. https://doi.org/10.3389/fpls.2016.00757
Boukar O, Belko N, Chamarthi S, Togola A, Batieno J, Owusu E, Haruna M, Diallo S, Umar ML, Olufajo O, Fatokun C (2018) Cowpea (Vigna unguiculata): genetics, genomics and breeding. Plant Breed. https://doi.org/10.1111/pbr.12589
Chen SL, Li YR, Cheng ZS, Liu JS (2009) GGE-biplot analysis of effects of planting density growth and yield components of high oil peanut. Acta Agron Sin 35:1328–1335
Cravero V, Esposito MA, Anido FL, Garcia SM, Cointry E (2010) Identificación of an ideal test environment for asparagus evaluation by GGE-biplot analysis. Aust J Crop Sci 4(4):273–277
de Oliveira DSV, Franco LJD, de Menezes-Júnior JA, Damasceno-Silva KJ, de Moura RM, das Neves AC, de Sousa FS (2017) Adaptability and stability of the zinc density in cowpea genotypes through GGE-biplot method. Rev Cienc Agron 48:783–791. https://doi.org/10.5935/1806-6690.20170091
Gauch HG, Zobel RW (1996) AMMI analysis of yield trials. In: Kang MS, Gauch HG (eds) Genotype by environment interaction. CRC Press, Boca Raton, Florida, pp 85–122
Gedif M, Yigzaw D, Tsige G (2014) Genotype–environment interaction and correlation of some stability parameters of total starch yield in potato in Amhara region. Ethiopia J Plant Breed Crop Sci 6(3):31–40. https://doi.org/10.5897/JPBCS2013.0426
Gurmu F, Mohammed H, Alemaw G (2009) Genotype × environment interactions and stability of soybean for grain yield and nutrition quality. Afr Crop Sci J 17(2):87–99
Kamai N, Gworgwor NA, Wabekwa JW (2014) Varietal trials and physiological components determining yield differences among cowpea varieties in semiarid zone of Nigeria. ISRN Agron 2014:1–7. https://doi.org/10.1155/2014/925450
Kimbeng CA, Zhou MM, Da Silva JA (2009) Genotype x enviroment interactions and resources allocation in Suggarcane yield trials in the Rio Grande valley region of Texas. J Am Soc Suggar Cane Technol 29:11–24
Kumar B, Hooda E, Hooda BK (2018) GGE biplot analysis of multi-environment yield trials for wheat in Northern India. Adv Res 16:1–9. https://doi.org/10.9734/air/2018/43488
Luo J, Zhang H, Deng Z, Xu L, Yuan Z, Que Y (2013) Analysis of yield and quality traits in suggarcane varieties (lines) with GGE-biplot. Acta Agonomic Sin 39(1):142–152
Luo JYB, Pan YH, Que MP, Zhang L, Grisham (2015) Biplot evaluation of test environments and identification of mega environment for sugarcane cultivars in China. Sci Rep 5:15505
Lin C, Bins MR (1991) Genetics properties of four types of stability parameters. Theory Appl Genet 82:505–509
Owusu EY, Akromah R, Denwar NN, Adjebeng-Danquah J, Kusi F, Haruna M (2018) Inheritance of early maturity in Some Cowpea (Vigna unguiculata (L.) Walp.) genotypes under rain fed conditions in Northern Ghana. Adv Agric. https://doi.org/10.1155/2018/8930259(article ID 8930259)
Pacheco A, Rodriguea F, Alvarado M, Lopez M, Crossa J, Burgueno J (2016) GEA-R (genotype × environment analyses with R for Windows) version 4.0. https://doi.org/10.3389/fpls.2014.00217
Padi FK (2004) Relationship between stress tolerance and grain yield stability in cowpea. J Agric Sci Cambr 142:143–153
Rakshit S, Ganapathy KN, Gomashe SS, Rathore A, Gorade RB et al (2012) GGE biplot analysis to evaluate genotype, environment and their interactions in sorghum multi-location data. Euphytica 185(3):465–479. https://doi.org/10.1007/s10681-012-0648-6
SAS Institute Inc (2011) Base SAS 93 procedures guide. SAS Institute Inc, Cary
Sbongeleni WD, Hussein SSR, Admire ITS (2019) Genotype-by-region interactions of released sugarcane varieties for cane yield in the South African sugar industry. J Crop Improv. https://doi.org/10.1080/15427528.2019.1621974
Singh BB, Mohan DR, Dashiell KE, Jackai LEN (2002) Co-publication of International Institute of Tropical Agriculture (IITA) and Japan International Research Centre for Agricultural Sciences (JIRCAS0). IITA, Ibadan
Singh BB (ed) (2020) Cowpea: the food legume of the 21st century, vol 164. Wiley, New York
Sousa MBE, Damasceno-Silva KJ, Rocha MDM (2018) Genotype by environment interaction in cowpea lines using GGE biplot method. Rev Caatinga 31:64–71. https://doi.org/10.1590/1983-21252018v31n108rc
Suwarto D (2010) Genotype × environment interaction on rice Fe content. Gadjah Mada University, Yogyakarta
Yan W, Hunt LA, Sheng QL, Szlavnics Z (2000) Cultivar evaluation and mega-environment investigation based on GGE biplot. Crop Sci 40:596–605
Yan WK (2002) Singular-value partitioning in biplot analysis of multi-environment trial data. Agron J 94:990–996
Yan WK (2010) Optimal use of biplots in analysis of multi-location variety test data. Acta Agronomy Sinica 36:1805–1819
Yan W, Tinker NA (2006) Biplot analysis of multi-environment trial data: Principles and applications. Can J Plant Sci 86(3):623–645. https://doi.org/10.4141/P05-169
Yan W, Kang MS, Ma BL, Woods S, Cornelius PL (2007) GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci 47:643–655
Yan W, Holland JB (2010) A heritability-adjusted GGE biplot for test environment evaluation. Euphytica 171:355–369
Zhang PP, Song H, Ke XW (2016) GGE biplot analysis of yield stability and test location representativeness in proso millet (Panicum miliaceum L.) genotypes. J Integr Agric 15:1218–1227. https://doi.org/10.1016/S2095-3119(15)61157-1
Zhou CJ, Tian ZY, Li JY (2011) GGE-biplot analysis on yield stability and testing-site representativeness of soybean lines in multi-environment trials. Soybean Sci 30:318–322
Acknowledgements
Authors are very grateful to the Tropical Legumes III project of Bill and Melinda Gates Foundation for the financial support. We also express our heartfelt gratitude to Prof. Rajeev Kumar Varshney, Dr. Emmanuel Monyo and Dr. Chris Ojiewo for their contributions to this study. Finally, we thank the management of International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India, and International Institute of Tropical Agriculture (IITA), Nigeria for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Rights and permissions
About this article
Cite this article
Owusu, E.Y., Amegbor, I.K., Mohammed, H. et al. Genotype × environment interactions of yield of cowpea (Vigna unguiculata (L.) Walp) inbred lines in the Guinea and Sudan Savanna ecologies of Ghana. J. Crop Sci. Biotechnol. 23, 453–460 (2020). https://doi.org/10.1007/s12892-020-00054-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-020-00054-5