Abstract
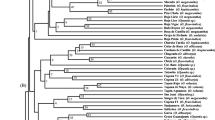
The genetic divergence among 34 genotypes belonging to 12 species of genus Cajanus were carried out using plant pest interaction and DNA marker analysis. Principal component analysis based on average percentage of pod damage caused by pod borer, plume moth, and blue butterfly in the field conditions, and growth of their larva and pupa on an artificial diet in vitro dispersed these genotypes into four coordinates evincing high genetic divergence as expected. DNA marker analysis using 11 pairs of SSR and nine ISSR primers showed higher polymorphism at the species level, and these primers exhibited variation with regard to average band informativeness, resolving power, and PIC value. No single primer was able to distinguish between all the 34 genotypes of Cajanus but nine species specific amplified fragments were generated by five ISSR primers. The pairwise Jaccard’s similarity coefficient and Nei’s genetic distance values revealed a higher level of inter-specific genetic variation in the genus Cajanus. The clustering of genotypes based on Jaccard’s similarity coefficient vis-a-vis Nei’s genetic distance agreed with the sectional classification of the genus Cajanus. Seven cultivars of C. cajan and the genotypes of their wild progenitor C. cajanifolius remained in one cluster, whereas accessions of C. platycarpus and C. scarabaeoides were out grouped. The rest of the genotypes belonging to nine species of Cajanus formed another cluster. The principal coordinate analysis also supported this clustering pattern. Moreover, these findings have good many implications for future breeding endeavors aimed at the introgression of pod borer resistance alleles.
Similar content being viewed by others
References
Agbagwa IO, Patil P, Datta S, Nadarajan N. 2014. Comparative efficiency of SRAP, SSR and AFLP-RGA markers in resolving genetic diversity in pigeonpea (Cajanus sp.). Indian J. Biotechnol. 13: 486–495
Armes NJ, Bond GS, Cooter RJ. 1992. The laboratory culture and development of Helicoverpa armigera. Natural Resources Institute, Bulletin No. 57. Natural Resources Institute, Chatham
Aruna R, Rao DM, Sivaramakrishnan S, Reddy LJ, Bramel B, Upadhyaya HD. 2008. Efficiency of three DNA markers in revealing genetic variation among wild Cajanus species. Plant Genet. Resour. 7: 113–121
Aruna R, Rao M, Reddy LJ, Upadhyaya HD, Sharma HC. 2005. Inheritance of trichomes and resistance to pod borer (Helicoverpa armigera) and their association in interspecific crosses between cultivated pigeonpea (Cajanus cajan) and its wild relative C. scarabaeoides. Euphytica 145: 247–257
Choudhary AK, Raje RS, Datta S, Sultana R, Ontagodi T. 2013. Conventional and Molecular Approaches towards Genetic Improvement in Pigeonpea for Insects Resistance. Am. J. Plant Sci. 4: 372–385
Choudhary AK, Vijayakumar AG. 2012. Glossary of Plant Breeding, A Perspective. LAP LAMBERT Academic Publishing
Saarbrücken Datta S, Kaashyap M, Kumar S. 2010. Amplification of chickpea-specific SSR primers in Cajanus species and their validity in diversity analysis. Plant Breed. 129: 334–340
Datta S, Singh P, Mahfooz S, Patil GP, Chaudhary AK, Agbagwa IO, Nadarajan N. 2013. Novel genic microsatellite markers from Cajanus scarabaeoides and their comparative efficiency in revealing genetic diversity in pigeonpea. J. Genet. 92: e24–e30
Dutta S, Kumawat G, Singh BP, Gupta DK, Singh S, et al. 2011. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol. 11: 17
Dutta S, Mahato AK, Sharma P, Raje RS, Sharma TR, Singh NK. 2013. Highly variable ‘Arhar’ simple sequence repeat markers for molecular diversity and phylogenetic studies in pigeonpea [Cajanus cajan (L.) Millisp.]. Plant Breed. 132: 191–196
Ganapathy KN, Gnanesh BN, Gowda MB, Venkatesha SC, Gomashe SS, Channamallikarjuna V. 2011. AFLP analysis in pigeonpea (Cajanus cajan (L.) Millsp.) revealed close relationship of cultivated genotypes with some of its wild relatives. Genet. Resour. Crop Evol. 58. 837–847
Green PWC, Stevenson PC, Simmonds MSJ, Sharma HC. 2003. Phenolic compounds on the pod surface of pigeonpea, Cajanus cajan, mediate feeding behaviour of larvae Helicoverpa armigera. J. Chem. Ecol. 29: 811–821
Grover A, Pental D. 2003. Breeding objectives and requirements for producing transgenics for major field crops of India. Curr. Sci. 84: 310–320
Kassa MT, Penmetsa RV, Carrasquilla-Garcia N, Sarma BK, Datta S, Upadhyaya HD, Varshney RK, Von Wettberg EJB, Cook DR. 2012. Genetic patterns of domestication in pigeonpea (Cajanus cajan (L.) Millsp.) and wild Cajanus relatives. PLoS One 7: e39563
Kumari DA, Reddy DJ, Sharma HC. 2006. Antixenosis mechanism of resistance in pigeonpea to the pod borer, Helicoverpa armigera. J. Appl. Entomol. 130: 10–14
Lewontin R. 1972. The Apportionment of Human Diversity. Evol. Biol. 6: 381–398
Mallikarjuna N, Saxena KB, Jadhav DR. 2011. Cajanus In: Kole C (ed.), Wild Crop Relatives: Genomic and Breeding Resources, Legume Crops and Forages, Springer-Verlag, Berlin, Heidelberg, pp 21–33
Mantel NA. 1967. The detection of disease clustering gene and a generalized regression approach. Cancer Res. 27: 209–220
Mishra RR. 2014. Molecular Mapping of genes conferring resistance to pod borer and pod fly in Cajanus cajan (L.) Millsp. Ph.D. Dissertation, Utkal University, Bhubaneswar, India
Mishra RR, Behera B, Panigrahi J. 2013a. Elucidation of Genetic relationships in the genus Cajanus using Random Amplified Polymorphic DNA marker analysis. J. Phylogenetics Evol. Biol. 1: 100–107
Mishra RR, Nag A, Panigrahi J. 2015. Development of dominant sequence characterized amplified region (SCAR) marker linked with plume moth (Exelastis atomosa Walsingham 1886) resistance in pigeon-pea. Chilean J. Agr. Res. 75: 497–501
Mishra RR, Sahu AR, Rath SC, Behera B, Panigrahi J. 2013b. Molecular mapping of locus controlling resistance to Helicoverpa armigera (Hubner) in Cajanus cajan L. (Millspaugh) using interspecific F2 mapping population. Nucleus 56: 91–97
Mishra RR, Sahu AR, Rath SC, Mishra SP, Panigrahi J. 2012. Cyto-morphological and molecular characterization of Cajanus cajan × C. scarabaeoides F1 hybrid. Nucleus 55: 27–35
Mohammadi SA, Prasanna BM. 2003. Analysis of genetic diversity in crop plants salient statistical tools and considerations. Crop Sci. 43: 1235–1248
Nadimpalli RJ, Jarret RL, Pathak SC, Kochert G. 1993. Phylogenetic relationships of the Pigeonpea (C. cajan) based on nuclear restriction fragment length polymorphisms. Genome 36: 216–223
Nei M. 1972. Genetic distance between populations. The American Naturalist. 106: 283–292
Nei M. 1973. Analysis of Gene Diversity in Subdivided Populations. Proc. Natl. Acad. Sci. USA 70: 3321–3323
Odeny DA, Jayashree B, Ferguson M, Hoisington D, Crouch J, Gebhardt C. 2007. Development, characterization and utilization of microsatellite markers in pigeonpea. Plant Breed. 126: 130–136
Panguluri SK, Janaiah J, Govil JN, Kumar PA, Sharma PC. 2006. AFLP fingerprinting in pigeonpea (Cajanus cajan L. Millsp.) and its wild relatives. Genet. Res. Crop Evol. 53: 523–531
Panigrahi J, Kumar DR, Mishra M, Mishra RP, Jena P. 2007. Genomic relationships in the genus Cajanus as revealed by seed protein (albumin and globulin) polymorphisms. Plant Biotechnol. Rep. 1: 109–116
Panigrahi J, Mishra RR, Sahu AR, Rath SC, Kole C. 2013. Marker-assisted breeding for stress resistance in crop plants. In: GR Rout, AB Das, eds., Molecular Stress Physiology of Plants, Springer, India, 387–426
Panigrahi J, Sahu AR, Mishra RR. 2015. Development and applications of DNA markers in pigeonpea: The present status and Future prospects. In: K Taski-Ajdukovic, SG Pandalai, eds., Applications of molecular markers in plant genome analysis and breeding, Research Signpost, Ontario, Canada, 91–117
Parde VD, Sharma HC, Kachole MS. 2012. Protease inhibitors in wild relatives of pigeonpea against the cotton bollworm/legume pod borer, Helicoverpa armigera. Am. J. Plant Sci. 3: 627–635
Patil PG, Datta S, Agbawa IO, Singh IP, Soren KR, Das A, Choudhary AK, Chaturvedi SK. 2014. Using AFLP-RGA markers to assess genetic diversity among pigeonpea (Cajanus cajan) genotypes in relation to major diseases. Acta Bot. Bras. 28: 198–205
Prevost A, Wilkinson MJ. 1999. A new system of comprising PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 98. 107–112
Pundir RPS, Singh RB. 1985. Crossability relationships among Cajanus, Atylosia and Rhynchosia species and detection of crossing barriers. Euphytica 34: 303–308
Ratnaparkhe MB, Gupta VS. 2007. Pigeonpea In: C Kole. ed., Genome Mapping and Molecular Breeding in Plants, Volume 3: Pulses, Sugar and Tuber Crops, Springer-Verlag Berlin Heidelberg, pp 133–145
Ratnaparkhe MB, Gupta VS, Ven Murthy MR, Ranjekar PK. 1995. Genetic fingerprinting of pigeonpea (Cajanus cajan (L.) Millsp) and its wild relatives using RAPD markers. Theor. Appl. Genet. 91: 893–898
Reed W, Lateef SS. 1990. Pigeonpea: Pest management. In: Nene YL, Hall SDand Sheila VK (eds.), The Pigeonpea. CAB International. Wallingford, UK, International Crops Research Institute For the Semi Arid Tropics, Patancheru, Andhra Pradesh, India, pp. 349–374
Rohlf FJ. 2000. NTSYSpc: Numerical Taxonomy and Multivariate Analysis System, Version 2.02. Exeter Software, Setauket, New York
Sahu AR. 2015. Construction of Genetic Map in Pigeonpea based on inter-specific F2 population using RAPD, SSR markers and simple inherited trait loci (SITLs). Ph.D. Dissertation, Sambalpur University, Odisha, India
Sahu AR, Mishra RR, Rath SC, Panigrahi J. 2015. Construction of interspecific molecular genetic map of pigeonpea using SCoT, RAPD, ISSR markers and SITLs. Nucleus 58: 23–31
Saxena KB. 2008. Genetic improvement of pigeonpea-A review. Tropical Plant Biol. 1: 159–178
Saxena KB, Kumar RV, Rao PV. 2002. Pigeonpea nutrition and its improvement. J. Crop Prod. 5: 227–260
Saxena RK, Prathima C, Saxena KB, Hoisington DA, Singh NK, Varshney RK. 2010b. Novel SSR markers for polymorphism detection in pigeonpea (Cajanus spp.). Plant Breed. 129: 142–148
Saxena RK, Saxena KB, Kumar RV, Hoisington DA, Varshney RK. 2010a. Simple sequence repeat-based diversity in elite pigeonpea genotypes for developing mapping populations to map resistance to Fusarium wilt and sterility mosaic disease. Plant Breed. 129: 135–141
Saxena RK, von Wettberg E, Upadhyaya HD, Sanchez V, Songok S, Saxena KB, Kimurto PK, Varshney RK. 2014. Genetic Diversity and Demographic History of Cajanus spp. Illustrated from Genome-Wide SNPs. PLoS One 9: e88568
Sharma HC, Sujana G, Manohar Rao D. 2009. Morphological and chemical components of resistance to pod borer, Helicoverpa armigera in wild relatives of pigeonpea. Arth-Plant Int. 3:151–161
Singh AK, Rai VP, Chand R, Singh RP, Singh MN. 2013. Genetic diversity studies and identification of SSR markers associated with Fusarium wilt (Fusarium udum) resistance in cultivated pigeonpea (Cajanus cajan). J. Genet. 92: e24–e30
Sivaramakrishnan S, Seetha K, Rao AN, Singh L. 1997. RFLP analysis of cytoplasmic male sterile lines in pigeonpea (Cajanus cajan L. Millsp.). Euphytica 93: 307–312
Slansky FJ. 1982. Insect nutrition: an adaptationist’s perspective. Fla. Entomol. 65. 46–71
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J. 1997. An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparison with data from RFLPs and pedigree. Theor. Appl. Genet. 95. 163–173
Sneath PHA, Sokal RR. 1973. Numerical Taxonomy: The principles and practice of numerical classification
Freeman WH, Sujana G, Sharma HC, Manohar Rao D. 2008. Antixenosis and antibiosis components of resistance to pod borer, Helicoverpa armigera in wild relatives of pigeonpea. Int. J. Trop. Insect Sci. 28: 191–200
Upadhyaya HD, Reddy KN, Singh S, Gowda CLL. 2013. Pigeonpea diversity in Cajanus species and identification of promising sources for agronomic traits and seed protein content. Genet. Resour. Crop Evol. 60: 639–659
van der Maesen LJG. 1980. India is the native home of pigeonpea. In: JC Arends, G Boelema, CT de Groot, AJM Lee uwenberg, eds., Liber Gratulatorius in honorem HCD de Wit. Landbouwhoge School, Miscellaneous paper no 19.
Veenman H, Zonen BV, Wageningen, Netherlands, pp 257–262.
Cajanus DC. and Atylosia W. A. 2013. A revision of all taxa closely related to the pigeonpea, with notes on other related genera within the subtribe Cajaninae (En). Wageningen papers, 85-4. Agriculture University, Wageningen, pp 225
Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK,et al. 2012. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 30: 83–89
Varshney RK, Murali Mohan S, Gaur PM, Gangarao NVPR, Pandey MK, et al. 2013. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol. Adv. 31: 1120–1134
Varshney RK, Penmetsa RV, Dutta S, Kulwal PL, Saxena RK, et al. 2010. Pigeonpea genomics initiative (PGI): an international effort to improve crop productivity of pigeonpea (Cajanus cajan L.). Mol. Breed. 26: 393–408
Verulkar SB, Singh DP, Bhattacharya AK. 1997. Inheritance of resistance to pod fly and pod borer in the interspecific cross of pigeonpea. Theor. Appl. Genet. 95: 506–508
Wasike S, Okori P, Rubaihayo PR. 2005. Genetic variability of Asian and African pigeonpea as revealed by AFLP. Afr. J. Biotechnol. 4: 1228–1233
Yang S, Pang W, Harper J, Carling J, Wenzl P, Huttner E, Zong X, Kilian A. 2006. Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology (DArT). Theor. Appl. Genet. 113: 585–595
Yeh FC, Yang R, Boyle T. 1999. Popgene version 1.32: Microsoft Windows-based freeware for population genetic analysis. University of Alberta, Edmonton
Yoshida M, Shanower TG. 2000. Helicoverpa armigera larval growth inhibition in artificial diet containing freezedried pigeonpea pod powder. J. Agr. Urban Entomol. 17: 37–41
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sahu, A.R., Mishra, R.R. & Panigrahi, J. Estimation of genetic diversity among 34 genotypes in the genus Cajanus with contrasting host response to the pod borer and its allied pests. J. Crop Sci. Biotechnol. 19, 17–28 (2016). https://doi.org/10.1007/s12892-015-0045-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-015-0045-5