Abstract
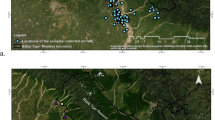
Feces of animals that forage on nectar and fruit, including many species of bats, often contain DNA that is low in quality and quantity. We developed an approach based on DNA from feces gathered passively to generate microsatellite data for individual lesser long-nosed bats (Leptonycteris yerbabuenae), which are important pollinators for columnar cacti and agave across much of Mexico and in the southwestern U.S. We collected feces from roosts near the U.S-Mexico border and developed a two-step amplification approach to characterize five highly polymorphic microsatellite loci from fecal DNA. Addition of a multiplex PCR step improved amplification success and conserved DNA extracts with a minimal increase in cost. In our initial screening of 433 samples, five focal loci distinguished individuals reliably, with a probability of identity (i.e., the probability of two unrelated individuals having the same microsatellite profile by chance) of 7.5E-09. Repeated analyses revealed a genotyping error rate < 2%. We explore the benefits and limits of our approach for population studies of lesser long-nosed bats and other nectivorous and frugivorous species that provide key ecosystem services and are often of conservation concern.




Similar content being viewed by others
References
Aizpurua O, Nyholm L, Morris E, Chaverri G, Herrera Montalvo LG, Flores-Martinez JJ, Lin A, Razgour O, Gilbert MTP, Alberdi A (2021) The role of the gut microbiota in the dietary niche expansion of fishing bats. Animal Microb 3:e76
Arandjelovic M, Guschanski K, Schubert G, Harris TR, Thalmann O, Siedel H, Vigilant L (2009) Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol Ecol Resour 9:28–36
Arnold AE, Andersen EM, Taylor MJ, Steidl RJ (2017) Using cytochrome b to identify nests and museum specimens of cryptic songbirds. Conserv Genet Resour 9:451–458
Arteaga MC, Medellín RA, Luna-Ortíz PA, Heady PA, Frick WF (2018) Genetic diversity distribution among seasonal colonies of a nectar-feeding bat (Leptonycteris yerbabuenae) in the Baja California Peninsula. Mamm Biol 92:78–85
Avena CV, Wegener Parfrey L, Leff JW, Archer HM, Frick WF, Langwig KE, Marm Kilpatrick A, Powers KE, Foster JT, McKenzie VJ (2016) Deconstructing the bat skin microbiome: influences of the host and the environment. Front Microbiol 7:e1753
Baldwin HJ, Hoggard SJ, Snoyman ST, Stow AJ, Brown C (2010) Non-invasive genetic sampling of faecal material and hair from the grey-headed flying-fox (Pteropus poliocephalus). Aust Mammal 32:56–61
Blanc L, Marboutin E, Gatti S, Gimenez O (2013) Abundance of rare and elusive species: empirical investigation of closed versus spatially explicit capture-recapture models with lynx as a case study. J Wildl Manag 77:372–378
Boston E, Montgomery I, Prodöhl PA (2009) Development and characterization of 11 polymorphic compound tetranucleotide microsatellite loci for the Leisler’s bat, Nyctalus leisleri (Vespertilionidae, Chiroptera). Conserv Genet 10:1501–1504
Brooks R, Williamson J, Hensley A, Butler E, Touchton G, Smith E (2003) Buccal cells as a source of DNA for comparative animal genomic analysis. Biotech Lett 25:451–454
Broquet T, Berset-Braendli L, Emaresi G, Fumagalli L (2007) Buccal swabs allow efficient and reliable microsatellite genotyping in amphibians. Conserv Genet 8:509–511
Ceballos G, Fleming TH, Chávez C, Nassar J (1997) Population dynamics of Leptonycteris curasoae (Chiroptera: Phyllostomidae) in Jalisco, Mexico. J Mammal 78:1220–1230
Cockrum LE (1991) Seasonal distribution of northwestern populations of the long-nosed bats, Leptonycteris sanborni family Phyllostomidae. Anales Del Instituto De Biología, Serie Zoología 62:181–202
Cook JD, Grant EHC, Coleman JTH, Sleeman JM, Runge MC (2021) Risks posed by SARS-CoV-2 to North American bats during winter fieldwork. Conserv Sci Pract 3:e410
Eggert LS, Eggert JA, Woodruff DS (2003) Estimating population sizes for elusive animals: the forest elephants of Kakum National Park, Ghana. Mol Ecol 12:1389–1402
Ferreira CM, Sabino-Marques H, Barbosa S, Costa P, Encarnação C, Alpizar-Jara R, Pita R, Beja P, Mira A, Searle JB, Paupério J, Alves PC (2018) Genetic non-invasive sampling (gNIS) as a cost-effective tool for monitoring elusive small mammals. Eur J Wildl Res 64:46
Fleming TH, Nunez RA, Sternberg LSL (1993) Seasonal changes in the diets of migrant and non-migrant nectarivorous bats as revealed by carbon stable isotope analysis. Oecologia 94:72–75
Fleming TH, Nelson AA, Dalton VM (1998) Roosting behavior of the lesser long-nosed bat, Leptonycteris curasoae. J Mammal 79:147–155
Fleming TH, Sahley CT, Holland JN, Nason JD, Hamrick JL (2001) Sonoran Desert columnar cacti and the evolution of generalized pollination systems. Ecol Monogr 71:511–530
Gaona O, Gómez-Acata ES, Cerqueda-García D, Neri-Barrios CX, Falcón LI (2019) Fecal microbiota of different reproductive stages of the central population of the lesser-long nosed bat, Leptonycteris yerbabuenae. PLoS ONE 14:1–19
Hedmark E, Ellegren H (2006) A test of the multiplex pre-amplification approach in microsatellite genotyping of wolverine faecal DNA. Conserv Genet 7:289–293
Hensley AH, Wilkins KT (1988) Leptonycteris nivalis. Mammalian Species 307:1–4
Ibouroi MT, Arnal V, Cheha A, Dhurham SAO, Montgelard C, Besnard A (2021) Noninvasive genetic sampling for flying foxes: a valuable method for monitoring demographic parameters. Ecosphere. https://doi.org/10.1002/ecs2.3327
Jones C, McShea WJ, Conroy MJ, Kunz TH (1996) Capturing mammals. In: Wilson DE, Cole FR, Nichols JD, Rudran R, Foster MS (eds) Measuring and monitoring biological diversity: standard methods for mammals. Smithsonian Press, Washington
Marrero P, Fregel R, Cabrera VM, Nogales M (2009) Extraction of high-quality host DNA from feces and regurgitated seeds: a useful tool for vertebrate ecological studies. Biol Res 42:147–151
Matschiner M, Salzburger W (2009) TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics 25:1982–1983
Mills LS, Citta JJ, Lair KP, Schwartz MK, Tallmon DA (2000) Estimating animal abundance using noninvasive DNA sampling: promise and pitfalls. Ecol Appl 10:283–294
Nagy M, Ustinova J, Mayer F (2010) Characterization of eight novel microsatellite markers for the neotropical bat Rhynchonycteris naso and cross-species utility. Conserv Genet Resour 2:41–43
Ober HK, Steidl RJ, Dalton VM (2005) Resource and spatial-use patterns of an endangered vertebrate pollinator, the lesser long-nosed bat. J Wildl Manag 69:1615–1622
O’Shea TJ, Bogan MA (2003) Monitoring trends in bat populations of the United States and territories: problems and prospects. U.S. Geological Survey, Biological Resources Discipline, Information and Technology Report, USGS/BRD/ITR-2003–00.
Oyler-McCance SJ, Fike JA, Lukacs PM, Sparks DW, O’Shea TJ, Whitaker JO (2018) Genetic mark-recapture improves estimates of maternity colony size for Indiana bats. J Fish Wildl Manag 9:25–35
Parejo-Farnés C, Albaladejo RG, Camacho C, Aparicio A (2018) From species to individuals: combining barcoding and microsatellite analyses from non-invasive samples in plant ecology studies. Plant Ecol 219:1151–1158
Peakall R, Smouse PE (2012) GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pfeiler SS, Conner MM, McKeever JS, Stephenson TR, German DW, Crowhurst RS, Prentice PR, Epps CW (2020) Costs and precision of fecal DNA mark–recapture versus traditional mark–resight. Wildl Soc Bull 44:531–542
Piggott MP, Taylor AC (2003) Remote collection of animal DNA and its applications in conservation management and understanding the population biology of rare and cryptic species. Wildl Res 30:1–13
Piggott MP, Bellemain E, Taberlet P, Taylor AC (2004) A multiplex pre-amplification method that significantly improves microsatellite amplification and error rates for faecal DNA in limiting conditions. Conserv Genet 5:417–420
Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE (2010) Global pollinator declines: trends, impacts and drivers. Trends Ecol Evol 25:345–353
Pourshoushtari RD, Ammerman LK (2021) Genetic variability and connectivity of the Mexican long-nosed bat between two distant roosts. J Mammal 102:204–219
Powell RA, Proulx G (2003) Trapping and marking terrestrial mammals for research: Integrating ethics, performance criteria, techniques, and common sense. ILAR J 44:259–276
Ramirez J (2011) Population genetic structure of the lesser long-nosed bat (Leptonycteris yerbabuenae) in Arizona and Mexico. Thesis, The University of Arizona, Tucson, USA
Ramirez J, Munguia-Vega A, Culver M (2011) Isolation of microsatellite loci from the lesser long-nosed bat (Leptonycteris yerbabuenae). Conserv Genet Resour 3:327–329
Ramírez-Francél LA, Garcia-Herreral LV, Losado-Prado S, Reinoso-Florez G, Sanchez-Hernandez A, Estrada-Villegas S, Lim BK, Guevara G (2022) Bats and their vital ecosystem services—a global review. Integr Zool 17:2–23
Ratto F, Simmons BI, Spake R, Zamora-Gutierrez V, MacDonald MA, Merriman JC, Tremlett CJ, Poppy GM, Peh KSH, Dicks LV (2018) Global importance of vertebrate pollinators for plant reproductive success: a meta-analysis. Front Ecol Environ 16:82–90
Regan EC, Santini L, Ingwall-King L, Hoffmann M, Rondinini C, Symes A, Taylor J, Butchart SHM (2015) Global trends in the status of bird and mammal pollinators. Conserv Lett 8:397–403
Roffler GH, Waite JN, Pilgrim KL, Zarn KE, Schwartz MK (2019) Estimating abundance of a cryptic social carnivore using spatially explicit capture–recapture. Wildl Soc Bull 43:31–41
Selkoe KA, Toonen RJ (2006) Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9:615–629
Shafer ABA, Wolf JBW, Alves PC, Bergström L, Bruford MW, Brännström I, Colling G, Dalén L, de Meester L, Ekblom R, Fawcett KD, Fior S, Hajibabaei M, Hill JA, Hoezel AR, Höglund J, Jensen EL, Krause J, Kristensen TN, Krützen M, McKay JK, Norman AJ, Ogden R, Österling EM, Ouborg NJ, Piccolo J, Popović D, Primmer CR, Reed FA, Roumet M, Salmona J, Schenekar T, Schwartz MK, Segelbacher G, Senn H, Thaulow J, Valtonen M, Veale A, Vergeer P, Vijay N, Vilà C, Weissensteiner M, Wennerström L, Wheat CW, Zieliński P (2015) Genomics and the challenging translation into conservation practice. Trends Ecol Evol 30:78–87
Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J (1996) Reliable genotyping from small DNA. Nucleic Acids Res 24:3189–3194
Taylor MJ, Mannan RW, U’Ren JM, Garber NP, Gallery RE, Arnold AE (2019) Age-related variation in the oral microbiome of urban Cooper’s hawks (Accipiter cooperii). BMC Microbiol 19:e47
US Fish and Wildlife Service (2016a) Species Status Assessment for the Lesser Long-Nosed Bat. Southwest Region, Albuquerque, NM.
US Fish and Wildlife Service (2016b) National white-nose syndrome decontamination protocol, version 04.12.2016. <https://www.fs.usda.gov/Internet/FSE_DOCUMENTS/stelprdb5378842.pdf. Accessed 20 Nov 1999
US Fish and Wildlife Service (2018) Endangered and threatened wildlife and plants; removal of the lesser long-nosed bat from the federal list of endangered and threatened wildlife. 50 CFR Part 17 [Docket No. FWS–R2–ES–2016–0138; FXES11130900000 178 FF09E42000] RIN 1018–BB91. Volume Fed. Regis.
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Waits LP, Paetkau D (2005) Noninvasive genetic sampling tools for wildlife biologists: a review of applications and recommendations for accurate data collection. J Wildl Manag 69:1419–1433
Waits LP, Luikart G, Taberlet P (2001) Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol 10:249–256
Walker JLS (2022) Phenology of lesser long-nosed bats and their food plants. Thesis, The University of Arizona, Tucson, USA
Walker FM, Williamson CHD, Sanchez DE, Sobek CJ, Chambers CL (2016) Species from feces: order-wide identification of Chiroptera from guano and other non-invasive genetic samples. PLoS ONE 11:1–22
Woodruff SP, Lukacs PM, Christianson D, Waits LP (2016) Estimating Sonoran pronghorn abundance and survival with fecal DNA and capture-recapture methods. Conserv Biol 30:1102–1111
Zylstra ER, Steidl RJ, Swann DE (2010) Evaluating survey methods for monitoring a rare vertebrate, the Sonoran desert tortoise. J Wildl Manag 74:1311–1318
Acknowledgements
We thank Melanie Bucci, Dave Dalton, Sherry Daugherty, Heatherlee Leary, Kara O’Brien, and Chelsey Trejo for field assistance; Suzanne Wilcox and Cristina Francois (Appleton-Whittell Research Ranch of the National Audubon Society) and Ami Pate, Danny Martin, Helen Fitting, and Jessica Garcia (National Park Service) for logistical support; Bob Behrstock, Karen LeMay, Scott Richardson, and Cecil and Carol Schwalbe for hosting capturing sessions at their homes; Christina Brown, Jamison Carey, Adam Leon, and Arielle de la Cruz for collecting fecal samples; Caroline Plecki, Nicolas Katz, and David Woods for assistance with data collection; Elizabeth Bowman, Alison Harrington, Nicole Colón-Carrión, and Shuzo Oita for laboratory support; and Barbara Fransway, Stacy Sotak, and Taylor Edwards of the University of Arizona Genetics Core for guidance in the laboratory. We are grateful to an anonymous reviewer for constructive feedback that helped to improve our manuscript.
Funding
We are grateful for financial support from the National Park Service’s Southwest Border Resource Protection Program, and from the School of Natural Resources and the Environment and School of Plant Sciences at the University of Arizona.
Author information
Authors and Affiliations
Contributions
RJS and AEA conceived of the study; JSW, RJS, AEA, and SAW collected samples; JSW and MML collected data; JSW and AEA analyzed data; JSW, AEA, and RJS wrote the manuscript, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors report no competing interests and have no conflicts of interest to report.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Walker, JL.S., Steidl, R.J., Wolf, S.A. et al. Improved amplification of fecal DNA supports non-invasive microsatellite genotyping of lesser long-nosed bats (Leptonycteris yerbabuenae). Conservation Genet Resour 16, 159–171 (2024). https://doi.org/10.1007/s12686-023-01344-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-023-01344-0