Abstract
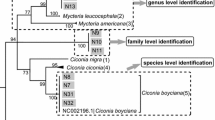
Bat fatality monitoring at wind turbines depends upon reliable identification of carcasses. Using reference mitochondrial cytochrome c oxidase I gene sequences mined from GENBANK and new sequences from collected samples, we constructed maximum likelihood trees including all 47 bat species found in the USA and tested the use of this locus for DNA barcoding these bat species. In this study, 80 % of species examined had distinct barcodes, including species currently listed as threatened or endangered. Nine of 17 Myotis bat species examined did not form distinct clades and had very low inter-specific genetic distances (1.4 %), thereby making this barcoding technique unreliable for some members of this genus. We then applied this technique to DNA samples from 892 bats salvaged from wind farms across four states. Using DNA barcoding, we were able to identify 14 carcasses to species that could not be identified in the field due to extensive decomposition and scavenging, and determined that another 18 carcasses had been misidentified in the field. Furthermore, we found field misidentifications increased with time until discovery. We conclude that DNA barcoding can improve the identification of salvaged bat carcasses especially when rare and uncommon species are encountered. This technique has other practical applications, such as identifying remains from hibernacula (potentially including carcasses of unknown bats with white-nose syndrome) or identifying species from fecal samples at roost sites or other locales.



Similar content being viewed by others
References
Ammerman LK, Hice CL, Schmidly DJ (2012) Bats of Texas. Texas A&M University Press, College Station
Arnett EB, Baerwald EF (2013) Impacts of wind energy development on bats: implications for conservation. In: Bat Evolution, Ecology, and Conservation. Adams RA, Pedersen SC eds. Springer Science + Business Media New York, pp 435-456
Arnett EB, Brown WK, Erickson WP, Fiedler JK, Hamilton BL, Henry TH, Jain A, Johnson GD, Kerns J, Koford RR, Nicholson CP, O’Connell TJ, Piorkowski MD, Tankersley RD (2008) Patterns of bat fatalities at wind energy facilities in North America. J Wildl Manag 72:61–78
Bennett VJ, Hale AM (2014) Red aviation lights on wind turbines do not increase bat-turbine collisions. Anim Conserv 17:354–358
Burland TM, Wilmer JW (2001) Seeing in the dark: molecular approaches to the study of bat populations. Biol Rev 76:389–409
Carstens BC, Dewey TA (2010) Species delimitation using a combined coalescent and information-theoretic approach: an example from North American Myotis bats. Syst Biol 59:400–414
Clare EL, Lim BK, Engstrom MD, Eger JL, Hebert PDN (2007) DNA barcoding of Neotropical bats: species identification and discovery within Guyana. Mol Ecol Notes 7:184–190
Clare EL, Lim BK, Fenton MB, Hebert PDN (2011) Neotropical bats: estimating species diversity with DNA barcodes. PLoS ONE 6(7):e22648. doi:10.1371/journal.pone.0022648
Cryan PM, Barclay RMR (2009) Causes of bat fatalities at wind turbines: hypotheses and predictions. J Mammal 90:1330–1340
Dewey TA (2006) Systematics and phylogeography of North American Myotis [dissertation]. Ann Arbor (MI): University of Michigan
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3:294–299
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp 41:95–98
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc B 270:313–321
Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN (2007) Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes 7:544–548
Kerr KCR, Stoeckle MY, Dove CJ, Weigt LA, Francis CM, Hebert PDN (2007) Comprehensive DNA barcode coverage of North American birds. Mol Ecol Notes 7:535–543
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Korstian JM, Hale AM, Bennett VJ, Williams DA (2013) Advances in sex determination in bats and its utility in wind-wildlife studies. Mol Ecol Resour 13:776–780
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V, Boehm JT, Machida RJ (2013) A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool 10:34
Magnacca KN, Brown MJF (2012) DNA barcoding a regional fauna: Irish solitary bees. Mol Ecol Resour 12:990–998
McCusker MR, Denti D, Van Geulpen L, Kenchington E, Bentzen P (2013) Barcoding Atlantic Canada’s commonly encountered marine fishes. Mol Ecol Resour 13:177–188
Nadin-Davis SA, Guerrero E, Knowles MK, Feng Y (2012) DNA barcoding facilitates bat species identification for improved surveillance of bat-associated rabies across Canada. Open Zool J 5:27–37
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Rodriguez RM, Ammerman LK (2004) Mitochondrial DNA divergence does not reflect morphological difference between Myotis californicus and Myotis ciliolabrum. J Mammal 85:842–851
Russell AL, Butchkoski CM, Saidak L, McCracken GF (2009) Road-killed bats, highway design, and the commuting ecology of bats. Endanger Species Res 8:49–60
Schmidly DJ (1991) The bats of Texas. Texas A&M University Press, College Station
Sequencher® version 5.0 sequence analysis software. Gene Codes Corporation, Ann Arbor, MI USA. http://www.genecodes.com
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Acknowledgments
This research was funded by NextEra Energy Resources; TCU researchers had unrestricted access to company data and complete independence in all aspects of data analysis, conclusions, and decision to publish the research. J.M.K. received a TCU Biology Department Adkins Fellowship. We thank Wolf Ridge Wind, LLC for logistical support and William Fleischer for help with molecular analyses. Thank you to the museums that contributed samples to this project: Angelo State Natural History Collection, Florida Museum of Natural History Museum of Southwestern Biology, and the Texas A&M University Teaching and Research Collection. Thank you to Faith Walker for providing us with early access to sequences for this project. We thank Jeff Gore and the Florida Fish and Wildlife Conservation Commission for contributing tissue samples. We also thank the many field technicians who participated in fatality searches at Wolf Ridge. We thank WEST, Inc. and the Minnesota Department of Commerce for providing samples from Minnesota, Colorado, West Virginia, and south Texas.
Author contributions
D.A.W., A.M.H. and V.J.B. developed the research project. J.M.K. conducted the molecular and taxonomic analyses. A.M.H. and V.J.B. provided tissue samples and funding. All authors contributed to writing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Korstian, J.M., Hale, A.M., Bennett, V.J. et al. Using DNA barcoding to improve bat carcass identification at wind farms in the United States. Conservation Genet Resour 8, 27–34 (2016). https://doi.org/10.1007/s12686-015-0509-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-015-0509-4