Abstract
The long noncoding DANCR functions as a tumor oncogene in many cancers, including colorectal cancer (CRC). However, the molecular mechanism of DANCR in CRC has not been explored. This study probed the function and potential mechanism by which DANCR contributes to the progression of CRC. The obtained data indicated that DANCR is overexpressed in CRC tissues and cell lines. Knockdown of DANCR hindered CRC cell proliferation, which was mediated by cyclin D1 and CDK4. Bioinformatic analysis, luciferase reporter assays and subcellular fractionation verified that DANCR directly binds to miR-508-5p. Moreover, DANCR acts as a miR-508-5p ceRNA to regulate expression of ATF1. In addition, upregulation of DANCR is attributed to H3K27 acetylation at the promoter region. In conclusion, our study confirmed that activation of lncRNA DANCR by H3K27 acetylation has an oncogenic role in CRC progression and provides a potential therapeutic target for CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Colorectal cancer (CRC) is the third most common tumor in the world and also the fourth most common cause of death [1]. CRC is the result of the combined effect of heredity and the environment [2]. Although much experience has accumulated in the treatment of CRC, survival based on drug resistance and metastasis is still a challenge [3]. To cope with these difficult clinical problems, it is particularly important to find effective and safe new molecules to diagnose and treat CRC.
Studies have shown that nonencoded RNA can be used as an important marker for the potential diagnosis, treatment and prognosis of tumors [4]. Long noncoding RNAs (lncRNAs) are nonprotein-coding RNAs that are more than 200 nucleotides in length and function by interacting with genes and proteins. As reported, lncRNAs participate in various cellular processes in cancer cells [5]. Indeed, lncRNAs perform numerous biological functions, such as proliferation, invasion and autophagy [6]. MicroRNAs (miRNAs) are 18- to 25-nucleotide noncoding RNAs that modulate expression of mRNAs [7]. LncRNAs compete with miRNAs to reduce the binding of miRNAs and mRNAs, which further affects expression of target genes [8].
It was recently found that differentiation antagonizing non-protein coding RNA (DANCR) is aberrantly expressed in different tumors, including CRC [9,10,11]. It has also been reported that roles of DANCR in the carcinogenesis, with an especial emphasis on its role in the development of osteosarcoma and lung, liver, pancreatic and CRC [11]. Study also showed DANCR may potentially provide a promising future therapeutic strategy for breast cancer treatment [12]. Previous research has reported that DANCR played a critical role in Doxorubicin-induced apoptosis in colorectal cancer cells through regulating the expression of MALAT1 [13]. Overall, although DANCR has been featured in several studies of CRC, the specific mechanism of DANCR still needs to be further studied. Moreover, to the best of our knowledge, there has been no research to date reporting the mechanism underlying the relationship between DANCR and ATF1 in CRC. In this study, DANCR was assessed in CRC tissues and cell lines. Moreover, the effects of DANCR on malignant behavior were detected through functional experiments, and the potential mechanisms of DANCR in CRC are herein discussed in depth. This study may provide new directions for therapeutic strategies for CRC.
2 Materials and methods
2.1 Patients and tissue samples of CRC
Sixty-nine fresh frozen CRC tissue and adjacent nontumor tissue samples (sampled at more than 5 cm from the tumor) were collected from The Affiliated Hospital of Qingdao University and stored at − 80 °C for this study. All patients signed the informed consent form for research purposes. The Hospital Ethics Review Committee approved the procedures of this study.
2.2 Cell lines and culture conditions
Human normal cell lines (NCM460) and human CRC cell lines (SW480, DLD-1, RKO, LOVO and HCT-116) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured as follows: in Roswell Park Memorial Institute (RPMI; Gibco, Grand Island, NY, USA) 1640 containing 10% FBS with 5% CO2 at 37 °C. C646 (Selleck Chemicals, Houston, TX, USA) was used at 10 μmol/L for 48 h, as needed.
2.3 RNA extraction and real-time quantitative polymerase chain reaction (qPCR)
Total RNA from CRC tissues and cell lines was extracted using TRIzol reagent (Invitrogen, Grand Island, NY, USA); mRNA was reverse transcribed to cDNA using PrimeScript RT Reagent Kit (Takara, Dalian, China). The DANCR expression level was measured using a SYBR Green PCR Master mix kit (Takara, Dalian, China). The sequences of the DANCR primers used were as follows: forward, AGT TCT GAC CAC GAG CTT TTC; reverse, GGT GCT ATG AGA TTC CGA GTT C. GAPDH was employed as an endogenous control. The 2−ΔΔCt method was used to calculate the results. The other primers used are listed in Table S1.
2.4 Cell transfection
The two si-DANCR molecules (si#1 and si#2), miR-508-5p mimics and inhibitors were obtained from GenePharma (Shanghai, China). Cells were cultured to 70% confluence. Lipofectamine 3000 Transfection Reagent (Life Technologies, Grand Island, NY, USA) was applied for cell transfection according to the manufacturer’s protocol. The sequences of the primers used are shown in Table S1.
2.5 Dual-luciferase reporter plasmid transfection
RB-REPORT™ plasmids (DANCR-wnt and ATF1-wnt) and their mutation plasmids (DANCR-mut and ATF1-mut) were obtained from RiboBio (Guangzhou, China). A Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA) was used to detect firefly luciferase and Renilla luciferase, and the activity of the former was normalized to that of the latter.
2.6 Cell counting kit-8 (CCK8) assay
A cell counting kit (CCK8 assay, Dojindo, Kumamoto, Japan) was used to measure the proliferation of CRC cells. A total of 2000 CRC cells were transfected with siRNAs and plated into 96-well plates for 24, 48, 72, and 96 h. A multifunctional microplate reader SpectraMax M5 (Sunnyvale, CA, USA) was used to measure absorbance at 490 nm.
2.7 In situ hybridization
DIG-labeled LNA-DANCR was designed and synthesized by RiboBio (Guangzhou, China). In brief, paraffin-embedded tissues were washed and permeabilized with Triton X-100 solution and then hybridized at 37 °C overnight. Diaminobenzidine solution (1:900; Boster Biological Technology) was used to determine expression. A BX51 microscope (Olympus Corporation) was used to observe staining intensity at a magnification of × 400.
2.8 Subcellular fractionation
A Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China) was used to extract and detect cytoplasmic and nuclear RNA. DANCR expression levels in the cytoplasmic and nuclear fractions were measured by qPCR. GAPDH and U6 were used as cytoplasmic and nuclear controls, respectively.
2.9 Chromatin immunoprecipitation assay (ChIP)
An EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was used to conduct ChIP. Briefly, tissues and cells were incubated with formaldehyde, and DNA‒protein crosslinks were generated for 20–30 min. Then, the crosslinked chromatin was sonicated into fragments. Both anti-H3K27ac (1:1,000; Abcam, Cambridge, UK) and anti-IgG (negative control) antibodies were used for immunoprecipitation. The precipitated DNA was analyzed using qPCR.
2.10 Protein extraction and western blotting
Total proteins were extracted from CRC tissues and cell lines with RIPA buffer (Thermo Fisher Scientific, Waltham, MA, USA). The protein lysates were separated by 10% SDS‒PAGE and transferred to PVDF membranes (Sigma), which were blocked with 5% skim milk for 2 h at room temperature. The membranes were incubated with primary antibodies (ATF-1 antibody (#25177; 1:1000) and β-actin antibody (#8457; 1:1000); Cell Signaling Technology, Beverly, MA, USA) overnight at 4 °C. Then, the membrane was incubated with HRP-labeled secondary antibody (Cell Signaling Technology, USA) for 1 h at room temperature. The protein bands were detected by an enhanced chemiluminescence kit (Millipore, Billerica, Massachusetts, USA).
2.11 Statistical analysis
Results are expressed as the mean ± standard deviation (SD) from six separate experiments. Comparisons between groups were analyzed using Student's test, and multiple group comparisons were analyzed using one‑way ANOVA with Tukey's post hoc test. Correlations between lncRNA and CRC clinical characteristics were determined using Pearson's chi‑squared test. All statistical analyses were performed using SPSS 22.0 (IBM Corp.). P < 0.05 was indicative of a significant difference.
3 Results
3.1 DANCR was overexpressed in CRC tissues and cells
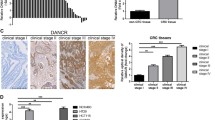
Based on the HCMDB dataset (http://hcmdb.i-sanger.com), we identified that DANCR was overexpressed in CRC tissue compared with matched adjacent normal tissue (n = 6; P < 0.05; Fig. 1A). Data from CRC tissue specimens also showed that the expression level of DANCR was higher in CRC tissue than in normal tissue (n = 6; P < 0.01; Fig. 1B). Additionally, using in situ hybridization with a DIG-labeled LNA-DANCR probe, we discovered that DANCR was predominantly located in the cytoplasm of CRC tissue and that its expression was higher than that in normal tissue (n = 6; P < 0.05; Fig. 1C and D). Furthermore, expression of DANCR, as measured by qPCR, was upregulated in CRC cell lines (SW480, DLD-1, RKO, LOVO and HCT-116; n = 6; P < 0.01; Fig. 1E). Among these CRC cells, LOVO and DLD-1 cells were much more abundant for the following study. To further identify the roles of DANCR in CRC cell behaviors, DANCR expression in both cell lines was silenced with two siRNAs (si#1 and si#2). Compared with that in the control group, the silencing efficiencies were significantly reduced (n = 6, P < 0.05; Fig. 1F). These results showed high expression of DANCR in CRC cells.
Abnormal DANCR expression in CRC and cell lines. A The HCMDB dataset showed the expression level of DANCR in CRC tissue (*P < 0.05). B Expression of DANCR was significantly elevated in CRC tissues. Statistical differences were analyzed using the Wilcoxon signed-rank test (*P < 0.01). C, D In situ hybridization of a DIG-labeled LNA- DANCR probe showed that DANCR was mainly distributed in the cytoplasm of CRC tissue. E Expression of DANCR in CRC cell lines (LOVO, HCT-116, DLD-1, SW480, and RKO) was detected by qPCR. F Knockdown efficiencies in cells transfected with si-DANCR. (n = 6; *P < 0.05 vs. NC)
3.2 Silencing of DANCR reduced proliferation in CRC cells
To investigate the effect of cell proliferation on the loss of DANCR function, we transfected both LOVO and DLD-1 cells with siRNAs. We found that the number of cells was significantly decreased compared with that in the control group, as evaluated by the CCK-8 assay (n = 6, P < 0.05; Fig. 2A and B). Meanwhile, several cell proliferation-related molecules [cyclin D1 and cyclin-dependent kinase 4 (CDK4)] were detected by qPCR assay, and the levels of cyclin D1 and CDK4 were downregulated after DANCR knockdown compared with the control group (n = 6, P < 0.05; Fig. 2C and D).
DANCR knockdown inhibits proliferation of CRC cells. A, B CCK-8 assays revealed that knockdown of DANCR inhibited proliferation in cells. B Histological analysis of rates of colony formation in the control (NC) and DANCR knockdown groups. C, D qPCR showed the mRNA expression level of cell cycle-related molecules [cyclin D1 and cyclin-dependent kinase 4 (CDK4)]. (n = 6; *P < 0.05 vs. NC)
3.3 DANCR sponges miR-508-5p, and its function was inhibited by miR-508-5p in CRC cells
A bioinformatics tool (https://starbase.sysu.edu.cn/ceRNA) was used to predict any downstream regulatory correlation for miRNA/DANCR. The results highlighted that miR-508-5p potentially binds to DANCR (Fig. 3A). Moreover, in the subcellular fractionation assay, DANCR was principally located in the cytoplasm of CRC cells (Fig. 3B). A luciferase reporter assay showed that the activity of DANCR-WT but not the DANCR-mut reporter construct was inhibited by miR-508-5p mimic overexpression (n = 6, P < 0.05; Fig. 3C). However, we noticed that upregulation of miR-508-5p expression reduced DANCR levels but that downregulation of miR-508-5p increased DANCR expression (n = 6, P > 0.05; Fig. 3D and E). These results indicated that miR-508-5p binds to DANCR. Furthermore, the roles of miR-508-5p in DANCR function in CRC cell proliferation were also detected by CCK8 assays. The reduced cell proliferation of CRC cells caused by DANCR knockdown was partly overcome by miR-508-5p inhibitor overexpression (n = 6, P < 0.05; Fig. 3F and G). These data strongly indicate that DANCR is partly mediated by miR-508-5p.
DANCR sponges miR-508-5p, and its function is suppressed by miR-508-5p. A Predicted miR-508-5p binding sites in the DANCR sequence. B Subcellular fractionation showed that DANCR was principally located in the cytoplasm of CRC cells. C The luciferase reporter assay showed that the luciferase activity of cells cotransfected with miR-508-5p and DANCR-WT reporter plasmids was decreased compared with that of the reporter with DANCR-mut or cotransfected miR-508-5p. D, E Cells were transfected with a miR-508-5p inhibitor or mimic, and DANCR expression was analyzed by qPCR. F, G CCK8 showed that the decreased number of live cells was partly upregulated in DANCR knockdown CRC cells. (n = 6; *P < 0.05 vs. NC; #P < 0.05 vs. si#1)
3.4 ATF1 was a target of miR-508-5p
To explore the molecular mechanism by which miR-508-5p promotes CRC progression, we used bioinformatics tools (http://www.targetscan.org/vert_72/) to predict the potential targets directly interacting with miR-508-5p. The data showed that ATF1 directly targets miR-508-5p (Fig. 4A). Through the luciferase reporter system, we observed that cotransfection of miR-508-5p mimics and ATF1-WT dramatically reduced luciferase activity but that it did not change when ATF1-MUT was transfected into cells (n = 6, P < 0.05; Fig. 4B). In addition, qPCR analysis showed that ATF1 was overexpressed by miR-508-5p inhibitor transfection but was decreased by transfection with the miR-508-5p mimic (n = 6, P < 0.05; Fig. 4C and D).
ATF1 is a target of miR-508-5p. A Bioinformatics tools found that ATF1 was a target of miR-508-5p. B The luciferase reporter system showed that after cotransfection of miR-508-5p mimics and ATF1-WT, luciferase activity was significantly reduced. C, D The results of by qPCR assays showed that the miR-508-5p mimic repressed expression of ATF1 in cells. (n = 6, *P < 0.05 vs. NC)
3.5 DANCR regulated ATF1 expression via miR-508-5p and proliferation in CRC cells
qPCR showed that while DANCR was upregulated in CRC tissues, ATF1 was also upregulated in the same tumor specimens, resulting in a significant positive correlation between DANCR and ATF1 [P < 0.01, R2 = 0.346; Fig. 5A]. The qPCR results showed that knockdown of DANCR downregulated expression of ATF1, which was partly rescued by the miR-508-5p inhibitor in CRC cells (n = 6, P < 0.05; Fig. 5B). Consistently, Western blotting showed that DANCR knockdown notably decreased ATF1 expression but that the miR-508-5p inhibitor partly rescued ATF1 expression in cells (n = 6, P < 0.05; Fig. 5C and D). Next, we analyzed the abnormal proliferation behavior caused by qPCR to determine whether DANCR is caused by the miR-508-5P target ATF1. After miR-508-5p inhibitor transfection, the decreased levels of CDK4 and cyclin D1 in DANCR-knockdown cells were partially recovered (n = 6, P < 0.05; Fig. 5E and F).
DANCR regulates ATF1 expression through miR-508-5p. A There was a significant positive correlation between DANCR and ATF1 (P < 0.01, R2 = 0.346). B qPCR results showed that knockdown of DANCR reduced expression of ATF1 but that the miR-508-5p inhibitor partly abolished this silencing effect. C, D Western blotting assays showed that ATF1 protein downregulation in DANCR knockdown cells was rescued by miR-508-5p inhibitor transfection. E, F qPCR analysis showed that the decreased levels of cyclin D1 and CDK4 in DANCR knockdown cells were partly rescued by miR-508-5p inhibitor transfection. (n = 6; *P < 0.05 vs. NC; #P < 0.05 vs. si#1)
3.6 H3K27 acetylation activated DANCR expression
The underlying mechanism of high DANCR levels in CRC cells needs to be investigated. Bioinformatics analysis (http://genome.ucsc.edu/) showed H3K27ac to be highly enriched in the DANCR promoter (Fig. 6A). To explore this, we used the anti-H3K27ac antibody of the ChIP assay in both CRC tissues and cancer cell lines. When compared with adjacent normal tissues, the expression level of H3K27ac at the DANCR promoter in CRC tissues was highly increased (n = 6, P < 0.05; Fig. 6B). Moreover, in CRC cells, levels of H3K27ac were enriched at the DANCR promoter (n = 6, P < 0.05; Fig. 6C). To assess whether H3K27ac is specific for DANCR transcription activation and expression upregulation, CRC cells were treated with C646 (a histone acetyltransferase inhibitor). Moreover, when compared with control cells, enrichment of H3K27ac was lower in CRC cells after C646 treatment (P < 0.05, Fig. 6D and E). We further found decreased expression of DANCR in C646-treated CRC cells (n = 6, P < 0.05; Fig. 6F).
DANCR is activated by H3K27 acetylation at the promoter region. A Bioinformatics analysis showing high enrichment of H3K27ac at the DANCR promoter. B ChIP assays detecting H3K27 acetylation at the DANCR promoter in CRC tissues. C ChIP assays detecting H3K27 acetylation at the DANCR promoter in CRC cells. D, E ChIP assays detecting H3K27 acetylation at the DANCR promoter after treatment of CRC cells with C646. F DANCR expression in C646-treated CRC cells using qPCR. (n = 6, *P < 0.05 vs. NC)
4 Discussion
Recent evidence has shown that lncRNAs have the ability to regulate complex and diverse biological functions, and they have important significance for different tumor types and for diagnosis, treatment and prognosis [14, 15]. In the last few years, multiple studies have established DANCR roles in promoting cancer tumor growth and migration among different cancers [16, 17]. DANCR is able to regulate hepatic malignant tumor progression via miR-125b-5p [18]. DANCR affects the resistance of gastric cancer by adjusting expression of MDR1 and MRP1 [19]. Studies have also shown the roles of DANCR in colorectal cancer tissues [20]. However, the specific mechanism through which DANCR exerts its biological functions has not yet been fully clarified. In particular, there are few studies on what causes the increase in DANCR in CRC. In this study, we found that DANCR was overexpressed, which is consistent with previous research [20]. We further found through CCK-8 experiments that DANCR knockdown can significantly inhibit the malignant proliferation of CRC cells. Studies have confirmed that CDK4 acts on cyclin D1 to further regulate cell proliferation [21,22,23]. The present study revealed the mechanisms of proliferation by which DANCR knockdown can reduce levels of cyclin D1 and CDK4 in cell lines. Although it would be helpful to examine the association of this gene with survival in colorectal cancer patients, samples from at least more than five years would be needed, and the samples used in this study were only obtained for at most one year. We will further evaluate DANCR and publish the survival association of this gene with colorectal cancer patients in the future.
Studies have shown that ceRNAs play an important role as lncRNAs that cause the development and progression of tumors [24]. However, in the cancer environment, in addition to lncRNA‒ceRNA signaling, many other factors might affect the occurrence and development of tumors. These factors might enhance the development of tumors or reduce the development of tumors. Such inconsistency requires further research. In this study, lncRNA‒ceRNA signaling was the focus of our research, and we found that DANCR was mainly expressed in the cytoplasm of CRC through in situ hybridization and subcellular fractionation assays. These results indicate that the ceRNA mechanism may exist and play an important regulatory role. DANCR, as a lncRNA, has many targets or targets, which might regulate CRC in different ways. Previous researches have reported that DANCR, as biomarkers for the diagnosis of colorectal cancer, might facilitate the growth and metastasis of CRC by regulating the epithelial-mesenchymal transition (EMT) process [14, 24]. Consistently, our results showed that miR-508-5p targets DANCR directly through bioinformatics analysis and dual-luciferase analysis. The results implied that DANCR acts as a sponge. Research has found that miR-508-5p plays a tumor-suppressor role in different cancers, including CRC [25, 26]. Moreover, our data showed that the effect of DANCR was partly reversed by miR-508-5p in CRC cells. Nevertheless, the patient’s genetic profile, primary tumor location and expression of DANCR could be insightful for future clinical practice in CRC therapy. This needs to be further investigated.
LncRNAs regulate the biological characteristics of cancer cells by competing with miRNAs for their shared strain [27]. In this study, we originally detected that miR-508-5p directly targets activating transcription factor 1 (ATF1). ATF1 participates in cell viability and cell transformation by adjusting the transcription of extracellular signals [28,29,30,31]. ATF1 plays a critical role in the proliferation and invasion of many cancer cells [32,33,34]. In this study, we found that ATF1 correlated positively with DANCR. Moreover, we validated that DANCR sponges miR-508-5p to regulate proliferation via ATF1. We also showed that a miR-508-5p inhibitor can partially inhibit the decrease in cycle-related molecules caused by DANCR knockdown. These studies have demonstrated that DANCR depletion-induced proliferation inhibition is partly mediated through ATF1 in CRC cells. According to bioinformatics website analysis (targetscan.com), whether in vivo or in vitro, a miRNA can have many target genes, and one target gene can correspond to multiple miRNAs. The series of in vitro experiments helps to verify our hypothesis. The in vivo experiments indicate relationships between miRNAs and target genes, at least partially. This needs to be further investigated.
Overall, the underlying mechanism causing DANCR overexpression in CRC cells needs to be further clarified. Previous studies have found that abnormal levels of lncRNAs are partly attributable to transcriptional activation regulated by acetylation [35,36,37,38]. In this study, the promoter region of DANCR showed that H3K27ac was obviously enriched in this region by bioinformatics analysis. Moreover, the level of H3K27ac at the DANCR promoter was highly increased and further upregulated DANCR expression in CRC tissue and cell lines. Histone acetyltransferases can control the histone acetylation process. In this study, we further used C646, a histone acetyltransferase inhibitor, to investigate the H3K27ac modification involved in DANCR expression. One study showed that C646 reduces H3K27ac enrichment at the DANCR promoter and causes downregulation of DANCR. These results indicate that the reason for DANCR upregulation is enrichment of H3K27ac at the DANCR promoter region. The limitation of this study was that other factors may function in the DANCR promoter region, such as methylation of H3K27 or lysine acetyltransferase 6A. We will further investigate to the association in CRC and publish the association in colorectal cancer patients in the future.
In summary, our study demonstrates that DANCR is activated by H3K27 acetylation and acts as an essential oncogene that promotes CRC proliferation. Mechanistically, DANCR functions as a miR-508-5p sponge to positively regulate ATF1 in CRC. This study reveals that DANCR is a potential therapeutic target in CRC.
Data availability
All the datasets generated and analyzed in the present study are included in this published article.
Abbreviations
- CRC:
-
Colorectal cancer
- lncRNA:
-
Long noncoding RNA
- miRNAs:
-
MicroRNAs
- ceRNAs:
-
Competing endogenous RNAs
- qPCR:
-
Real-time quantitative polymerase chain reaction
- siRNA:
-
Short interfering RNA
References
Tao J, Tu Y, Liu P, Tang Y, Wang F, Li Z, Li C, Li Y, Ma Y, Gu Y. Detection of colorectal cancer using a small molecular fluorescent probe targeted against c-Met. Talanta. 2021;226:122128.
Van Zutphen M, Boshuizen HC, Kenkhuis MF, Wesselink E, Geijsen A, De Wilt JHW, Van Halteren HK, Bilgen EJS, Keulen ETP, Janssen-Heijnen MLG, et al. Lifestyle after colorectal cancer diagnosis in relation to recurrence and all-cause mortality. Am J Clin Nutr. 2021;113:1447–57.
Chauhan S, Dhawan DK, Saini A, Preet S. Antimicrobial peptides against colorectal cancer-a focused review. Pharmacol Res. 2021;167:105529.
Karakas D, Ozpolat B. The role of LncRNAs in translation. Noncoding RNA. 2021;7:16.
Carelli S, Giallongo T, Rey F, Latorre E, Bordoni M, Mazzucchelli S, Gorio MC, Pansarasa O, Provenzani A, Cereda C, et al. HuR interacts with lincBRN1a and lincBRN1b during neuronal stem cells differentiation. RNA Biol. 2019;16:1471–85.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61.
Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14.
Cantile M, Di Bonito M, De Bellis MT, Botti G. Functional interaction among lncRNA HOTAIR and microRNAs in cancer and other human diseases. Cancers. 2021;13:570.
Jin SJ, Jin MZ, Xia BR, Jin WL. Long non-coding RNA DANCR as an emerging therapeutic target in human cancers. Front Oncol. 2019;9:1225.
Sun Y, Cao B, Zhou J. Roles of DANCR/microRNA-518a-3p/MDMA ceRNA network in the growth and malignant behaviors of colon cancer cells. BMC Cancer. 2020;20:434.
Ghafouri-Fard S, Khoshbakht T, Hussen BM, Baniahmad A, Taheri M, Samadian M. A review on the role of DANCR in the carcinogenesis. Cancer Cell Int. 2022;22:194.
Yan S, Teng L, Du J, Ji L, Xu P, Zhao W, Tao W. Long non-coding RNA DANCR aggravates breast cancer through the miR-34c/E2F1 feedback loop. Mol Med Rep. 2024;29:93.
Xiong M, Wu M, Peng D, Huang W, Chen Z, Ke H, Chen Z, Song W, Zhao Y, Xiang AP, Zhong X. LncRNA DANCR represses Doxorubicin-induced apoptosis through stabilizing MALAT1 expression in colorectal cancer cells. Cell Death Dis. 2021;12:24.
Bahreini F, Saidijam M, Mousivand Z, Najafi R, Afshar S. Assessment of lncRNA DANCR, miR-145-5p and NRAS axis as biomarkers for the diagnosis of colorectal cancer. Mol Biol Rep. 2021;48:3541–7.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6.
Lu Y, Hu Z, Mangala LS, Stine ZE, Hu X, Jiang D, Xiang Y, Zhang Y, Pradeep S, Rodriguez-Aguayo C, et al. MYC targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 levels. Cancer Res. 2018;78:64–74.
Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511.
Yang L, Jiang MN, Liu Y, Wu CQ, Liu H. Crosstalk between lncRNA DANCR and miR-125b-5p in HCC cell progression. Tumori. 2021;107:504–13.
Xu YD, Shang J, Li M, Zhang YY. LncRNA DANCR accelerates the development of multidrug resistance of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:2794–802.
Wang Y, Lu Z, Wang N, Feng J, Zhang J, Luan L, Zhao W, Zeng X. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50:1–17.
Zhou L, Liu R, Liang X, Zhang S, Bi W, Yang M, He Y, Jin J, Li S, Yang X, et al. lncRNA RP11-624L4.1 is associated with unfavorable prognosis and promotes proliferation via the CDK4/6-cyclin D1-Rb-E2F1 pathway in NPC. Mol Ther Nucleic Acids. 2020;22:1025–39.
Digiacomo G, Fumarola C, La Monica S, Bonelli MA, Cretella D, Alfieri R, Cavazzoni A, Galetti M, Bertolini P, Missale G, et al. Simultaneous combination of the CDK4/6 inhibitor palbociclib with regorafenib induces enhanced anti-tumor effects in hepatocarcinoma cell lines. Front Oncol. 2020;10:563249.
Hindupur SV, Schmid SC, Koch JA, Youssef A, Baur EM, Wang D, Horn T, Slotta-Huspenina J, Gschwend JE, Holm PS, et al. STAT3/5 inhibitors suppress proliferation in bladder cancer and enhance oncolytic adenovirus therapy. Int J Mol Sci. 2020;21:1106.
Wang B, Chen W, Zhao Z, Sun Y, Huang Y. LncRNA-DANCR promotes growth and metastasis of colorectal cancer via activating epithelial-mesenchymal transition process. Transl Cancer Res. 2019;8:2517–25.
Shang Y, Zhang Z, Liu Z, Feng B, Ren G, Li K, Zhou L, Sun Y, Li M, Zhou J, et al. miR-508-5p regulates multidrug resistance of gastric cancer by targeting ABCB1 and ZNRD1. Oncogene. 2014;33:3267–76.
Zhang C, Yao K, Zhang J, Wang C, Wang C, Qin C. Long noncoding RNA MALAT1 promotes colorectal cancer progression by acting as a ceRNA of miR-508-5p to regulate RAB14 expression. Biomed Res Int. 2020;2020:4157606.
Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234:10080–100.
Klemm DJ, Roesler WJ, Boras T, Colton LA, Felder K, Reusch JE. Insulin stimulates cAMP-response element binding protein activity in HepG2 and 3T3-L1 cell lines. J Biol Chem. 1998;273:917–23.
Liu K, Cho YY, Yao K, Nadas J, Kim DJ, Cho EJ, Lee MH, Pugliese A, Zhang J, Bode AM, et al. Eriodictyol inhibits RSK2-ATF1 signaling and suppresses EGF-induced neoplastic cell transformation. J Biol Chem. 2011;286:2057–66.
Jin XL, O’Neill C. The presence and activation of two essential transcription factors (cAMP response element-binding protein and cAMP-dependent transcription factor ATF1) in the two-cell mouse embryo. Biol Reprod. 2010;82:459–68.
Bleckmann SC, Blendy JA, Rudolph D, Monaghan AP, Schmid W, Schütz G. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol Cell Biol. 2002;22:1919–25.
Wei YQ, Guo YF, Yang SM, Ma HH, Li J. MiR-340-5p mitigates the proliferation and activation of fibroblast in lung fibrosis by targeting TGF-β/p38/ATF1 signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24:6252–61.
Guo Y, Sun W, Gong T, Chai Y, Wang J, Hui B, Li Y, Song L, Gao Y. miR-30a radiosensitizes non-small cell lung cancer by targeting ATF1 that is involved in the phosphorylation of ATM. Oncol Rep. 2017;37:1980–8.
Liang X, Zhou D, Wei C, Luo H, Liu J, Fu R, Cui S. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS ONE. 2012;7: e33861.
Zhang E, Han L, Yin D, He X, Hong L, Si X, Qiu M, Xu T, De W, Xu L, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–101.
Chen F, Qi S, Zhang X, Wu J, Yang X, Wang R. lncRNA PLAC2 activated by H3K27 acetylation promotes cell proliferation and invasion via the activation of Wnt/β-catenin pathway in oral squamous cell carcinoma. Int J Oncol. 2019;54:1183–94.
Gao Y, Luo X, Zhang J. LincRNA-ROR is activated by H3K27 acetylation and induces EMT in retinoblastoma by acting as a sponge of miR-32 to activate the Notch signaling pathway. Cancer Gene Ther. 2021;28:42–54.
Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, Chen X, Han M. Activation of LncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting microRNA-125b in breast cancer. Mol Cancer. 2019;18:3.
Acknowledgements
All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Natural Science Foundation of Shandong Province (2019SD02014).
Author information
Authors and Affiliations
Contributions
STD and HY conceived and designed the experiments. STD, HHT, LQ and CL performed the experiments and analyzed the data. STD and SMQ provided samples. STD and CL provided guidance. STD and HY wrote the paper.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All patients signed the informed consent form for research purposes. The Hospital Ethics Review Committee approved the procedures of this study, the ethics approval number: QDFY20221103867. The experiments on humans/human data/the use of human tissue samples in this study were performed in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Han, Y., Shan, TD., Huang, HT. et al. Activation of lncRNA DANCR by H3K27 acetylation regulates proliferation of colorectal cancer cells. Discov Onc 15, 249 (2024). https://doi.org/10.1007/s12672-024-01124-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-024-01124-8