Abstract
Cervical cancer (CC) is considered as the fourth most common women cancer globally.that shows malignant features of local infiltration and invasion into adjacent organs and tissues. There are several individual studies in the literature that explored CC-causing hub-genes (HubGs), however, we observed that their results are not so consistent. Therefore, the main objective of this study was to explore hub of the HubGs (hHubGs) that might be more representative CC-causing HubGs compare to the single study based HubGs. We reviewed 52 published articles and found 255 HubGs/studied-genes in total. Among them, we selected 10 HubGs (CDK1, CDK2, CHEK1, MKI67, TOP2A, BRCA1, PLK1, CCNA2, CCNB1, TYMS) as the hHubGs by the protein–protein interaction (PPI) network analysis. Then, we validated their differential expression patterns between CC and control samples through the GPEA database. The enrichment analysis of HubGs revealed some crucial CC-causing biological processes (BPs), molecular functions (MFs) and cellular components (CCs) by involving hHubGs. The gene regulatory network (GRN) analysis identified four TFs proteins and three miRNAs as the key transcriptional and post-transcriptional regulators of hHubGs. Then, we identified hHubGs-guided top-ranked FDA-approved 10 candidate drugs and validated them against the state-of-the-arts independent receptors by molecular docking analysis. Finally, we investigated the binding stability of the top-ranked three candidate drugs (Docetaxel, Temsirolimus, Paclitaxel) by using 100 ns MD-based MM-PBSA simulations and observed their stable performance. Therefore the finding of this study might be the useful resources for CC diagnosis and therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cancer has the largest clinical, social, and economic impact in terms of cause-specific disability-adjusted life years [1]. Cervical cancer (CC) is the fourth most common cancer in women and has the fourth highest mortality rate globally [2]. The most common causes of cancer-related mortality among individuals with CC are invasion and metastasis by CC cells, which are associated with a poor prognosis [3, 4]. Fortunately, effective primary and secondary preventive methods, including as human papillomavirus (HPV) vaccine and yearly cytology smears, are available for CC. Almost all CC patients have a long-term infection with high-risk HPV types [5,6,7]. For early-stage and low-risk CC, surgery, chemotherapy, or radiation have shown to be effective [8,9,10]. However, metastatic cervical cancer (mCC) has a 5-year survival rate of 16.5 percent [11]. Additionally, the efficacy of chemotherapy and radiation is reduced by their side effects. Therefore, exploration of biological causes as well as the development of new treatment targets and techniques for CC are required to increase patient survival.
The identification of biomarkers that might possibly alleviate the disorder's pathogenesis could be a motivating factor that leads to the development of more effective therapy options in the future [12]. The potential benefits of molecular biomarkers suggest that they might improve the efficacy of CC diagnostic and treatment. Bioinformatics techniques are now widely used in several of biological research areas. Recently, sequencing tools have become more widely available, allowing researchers to make significant findings in both computational biology and molecular therapies [13]. Protein–Protein Interactions (PPI) were previously utilized to identify the hub genes that may be responsible for the disease, and a co-expression network was employed to validate the listed genes using a heat map based on their co-regulation scores [14,15,16,17]. Protein 3D structures are important for the fields in evolutionary biology and biotechnology, such as protein function prediction and drug design [18].
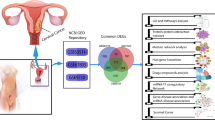
However, new drug development is a difficult, time-consuming, and costly endeavor. The major challenges are to identify disease-causing drug target proteins and drug agents that can alleviate disease severity by interacting with the target proteins. As opposed to developing a new drug, repurposing existing drugs for certain conditions might save time and money. By this time, numerous studies have proposed different sets of hub/key-genes to explore molecular mechanisms and pathogenetic processes in CC [19,20,21,22,23,24]. Some of these studies have also been suggested their hub-genes or studied-gene guided candidate drugs for the treatment of CC [23, 24]. By the literature review, we observed that their suggested CC-causing hub-genes (HubGs) or studied-genes sets as well as candidate drugs are not so consistent in different published articles. On the other hand, so far, none of these studies investigated the performance of their suggested drugs against the other published hub-genes mediated target proteins. Obviously, more representative hub-genes are required to explore more effective candidate drugs against CC. Therefore, in this study, our main objectives are to (1) explore hub of the HubGs (hHubGs) highlighting their functions, pathways, and regulatory factors, (2) explore hHubGs-guided candidate drugs for the treatment against CC, and (3) cross-validation of the proposed candidate drugs against the state-of-the-arts alternatives independent target proteins published by others. The workflow of this study is demonstrated in Fig. 1.
2 Materials and methods
In this study, the necessary meta-drug target and agents were collected from different online sources and published articles to explore globally most effective repurposable drugs for the treatment against CC by using the integrated bioinformatics approaches.
2.1 Metadata sources and descriptions
We have collected metadata for both drug targets and agents associated with CC to reach the goal of this study as described below.
2.2 Collection of studied/hub-DEGs to explore drug targets
Several research groups already published different sets of studied or hub differentially expressed genes (studied/hub-DEGs) associated with CC [12, 13, 20, 22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71]. Here DEGs indicates the differentially expressed genes between CC and control samples. The present study included the articles in 2010–2021 associated with CC infections.To select the top-ranked hub-genes (independent meta-receptors) associated with CC disease; we reviewed 52 published articles and collected 255 hub meta-receptors to explore key genes (KGs) by protein–protein interaction (PPI) network analysis [see Table S1]. The proteins corresponding to our proposed KGs would be considered as the key drug targets.
2.3 Collection of drug agents
Several research groups already suggested transcriptome-guided different sets of repurposable drugs for the treatment against CC [23, 24, 69, 70, 72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91]. We collected host transcriptome-guided 80 meta-drug agents by the literature review of CC disease [see Table S2] for exploring candidate drugs.
2.4 Protein–protein interaction (PPI) network analysis of all Hub/Studied-DEGs
The STRING online database (https://string-db.org/) was used to construct the PPI network of DEGs. [92]. We utilized the Cytoscape software to enhance the quality of the PPI network [93]. To select common Hub-Genes (cHubGs) or common Hub-Proteins (cHubPs) from the PPI network, the Cytoscape plugin cytoHubba was implemented [93, 94]. The PPI network is comprised of a number of nodes and edges that represent proteins and their interactions, respectively. A node with the largest number of significant interactions/connections/edges with other nodes is considered the top-ranked cHubGs. Four topological analyses of PPI network were applied to select the cHubGs (Degree [95], BottleNeck [96], Betweenness [97], and Stress [98]). The Cytoscape software's Molecular Complex Detection (MCODE) (http://apps.cytoscape.org/apps/mcode) plugin was used to identify the most significant modules in the PPIs network. MCODE clustering detected highly interconnected portions, which helps the study with effective drug design. By identifying densely connected regions, MCODE was utilized to represent molecular complexes in the PPIs network [99]. The hub of the hub-genes (hHubGs) were then selected since they were shared by both cHubGs and MCODE clustering genes.
2.5 Differential expression patterns analysis of hHubGs
To validate the expression of the hHubGs, the Gene Expression Profiling Interactive Analysis (GEPIA) website was applied to analyze the data of RNA sequencing expression based on thousands of samples from the GTEx projects and TCGA.
2.6 GO terms and KEGG pathway enrichment analysis of all Hub/Studied-DEGs
Gene ontology (GO) functional and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment/annotation/over-representation analysis [100, 101] is a widely used approach to determine the significantly annotated/enriched/over-represented functions/classes/terms and pathways by the identified KGs. It is indeed vital for understanding how genes work at the molecular level and what role they play in the cell. Biological Process, Cellular Component, and Molecular Function are the three categories of GO terms [102]. We used the DAVID online tool (https://david.ncifcrf.gov/tools.jsp) to perform GO and KEGG enrichment analysis [103]. The p-value 0.05 was selected as the significance level.
2.7 Regulatory network analysis of hHubGs
Using the NetworkAnalyst online tool, we performed TFs-hHubGs and miRNAs-hHubGs interaction network analysis on hHubGs to uncover major transcriptional regulatory transcription factors (TFs) and post-transcriptional regulatory micro-RNAs (miRNAs) [104]. The ENCODE (https://www.encodeproject.org/) [105] and RegNetwork repository [106] databases were used to construct the TFs- hHubGs and miRNAs- hHubGs interaction networks, respectively. To enhance the quality of the networks, the Cytoscape software [93] was utilized. Then, we determined the regulators (TFs or miRNAs) based on their high connectivity with hHubGs.
2.8 Drug repurposing by molecular docking study
We performed a molecular docking analysis of our suggested receptor proteins with drug agents to propose in-silico validated efficient candidate drugs for the treatment of CC. As previously mentioned in the data sources (see Table S1), we considered our proposed hHubGs based key proteins (KPs) and their regulatory key TFs proteins as drug target proteins and 77 meta-drug agents. Both receptor proteins and meta-drug agents require 3-Dimensional (3D) structures for molecular docking studies. All of the targeted proteins' 3D structures were downloaded from the Protein Data Bank (PDB) [107] and SWISS-MODEL [108]. All meta-drug agents' 3D structures were downloaded from the PubChem database [109]. Using discovery studio visualizer 2019 [110], the 3D structures of the target proteins were displayed, and target chains that were not part of the gene were deleted. The protonation state of protein was assigned using the PDB2PQR and H + + servers [111, 112]. As well, all absent hydrogen atoms were properly added. The pKa for the receptor amino acids were examined under the physical conditions of pH = 7, salinity = 0.15, external dielectric = 80, and internal dielectric = 10. Then, using AutoDock tools, the receptor was prepared for molecular docking study by eliminating water molecules, and ligand heteroatoms and by addition of polar hydrogens [113]. The grid box was generated over the entire surface of the proteins. The ligands were prepared for molecular docking study by using AutoDock tools to set the torsion tree and rotatable and nonrotatable bonds in the ligand. AutoDock Vina was used to calculate binding affinities between target proteins and drug agents [114]. The exhaustiveness parameter was set to 10. PyMol [115] and discovery studio visualizer 2019 [110] were used to analyse the docked complexes for surface complexes, types, and distances of non-covalent bonds. Let Aij denotes the binding affinity between ith target protein (i = 1, 2, …, m) and jth drug agent (j = 1, 2, …, n). To select the top-ranked lead compounds as the candidate drugs, we ordered the drug target proteins and agents according to the descending order of row sums \({\sum }_{j=1}^{n}{A}_{ij}\), j = 1,2,…,m, and column sums \({\sum }_{i=1}^{m}{A}_{ij}\), j = 1,2,…,n, respectively.
2.9 Molecular dynamic (MD) simulations
To discover the dynamic behavior of the top-ranked protein–ligand complexes, MD simulations were performed using YASARA Dynamics software [116], and the AMBER14 force field [117]. A total of six different systems were used to run MD simulation. The systems included Docetaxel-CDK1, Temsirolimus-CHEK1, Paclitaxel-TOP2A, apo-CDK1, apo-CHEK1, and apo-TOP2A.
For the complexes, the ligand parameters for the complexes were assigned using AutoSMILES [118] algorithms. A TIP3P [119] water model in a simulation cell was used for optimized and solvated the hydrogen bonding network of the protein–ligand complexes before the simulation. With a solvent density of 0.997 gL1, periodic boundary conditions were maintained. Titratable amino acids in the complexes were subjected to pKa calculation during solvation. The initial energy minimization process of each simulation system, consisting of of 55,410 ± 10, 72,287 ± 10, and 96,252 ± 10 atoms for CDK1_Docetaxel, CHEK1_Temsirolimus, and TOP2A_Paclitaxel complexes were performed by a simulated annealing method respectively, using the steepest gradient approach (5000 cycles). For the details of MD simulation methods see our previous paper [14, 15]. The trajectories were recorded every 250 ps for further analysis, and subsequent analysis was implemented by default script of YASARA [120] macro and SciDAVis software available at http://scidavis.sourceforge.net/. All snapshots were then subjected to YASARA software's MM-Poisson–Boltzmann surface area (MM-PBSA) binding free energy calculation using the formula below [121],
Here, MM-PBSA binding energy was calculated using YASARA built-in macros using AMBER 14 as a force field, with larger positive energies indicating better binding [122].
3 Results
3.1 Basic characteristics of the selected studies
The goal of this study was to identify the potential biomarkers and repurposing drug agents against cc. To screening the published articles, we conducted title, abstract, result, and conclusion with cervical cancer, hub genes/targets/receptors/proteins, number of drug agents/compounds/ligands, and collected 52 published articles from online databases to collect the hub genes or study genes. To select the top-ranked independent meta-receptors associated with CC disease, we gather 255 hub meta-receptors from 52 published articles to explore hub of the Hub-DEGs (hHubGs) by protein–protein interaction (PPI) network analysis [see Table S1]. On the other hand, we acquired 23 published papers from online databases to select the meta-drug agents/compounds/ligands against CC disease and accumulated host transcriptome-guided 77 meta-drug agents from 23 reputed published papers against CC disease [see Table S2] for exploring the candidate drugs.
3.2 Identification of hub of hub-genes (hHubGs)
To identify the hHub-proteins, the protein protein interaction (PPI) network analysis was utilized by using the all collected hub-DEGs from the selected published papers. The PPI network of cDEGs was constructed using STRING database (Fig. 2a) which contains 229 nodes and 4488 edges. We selected top-ranked sixteen (16) cHubGs {PLK1, TP53, GAPDH, VEGFA, CDK2, MKI67, CHEK1, CDK1, BRCA1, TOP2A, CLTC, CCNA2, JUN, CCND1, CCNB1, TYMS} applying four topological measures in the PPI network. Then, using MCODE, clusters were selected from the PPI network. It was shown that the most significant cluster had 27 nodes and 350 edges (see Table S3). MCODE analysis demonstrated that the most significant cluster contained the ten hHubGs {CDK2, CHEK1, MKI67, TOP2A, CDK1, BRCA1, PLK1, CCNA2, CCNB1, TYMS} (see Fig. 2b). These top hHubGs may be focused for the pre-clinical potential drug target molecule that may open a new era of therapeutic targets.
a Protein-proteins interaction network for common differentially expressed genes of CC, and edges specify the interconnection in the middle of two genes. The analyzed network holds 229 nodes and 4488 edges. Surrounding nodes (PLK1, TP53, GAPDH, VEGFA, CDK2, MKI67, CHEK1, CDK1, BRCA1, TOP2A, CLTC, CCNA2, JUN, CCND1, CCNB1, TYMS) represented the hub genes. b Module analysis network obtained from MCODE analysis. Surrounding nodes (CDK2, CHEK1, MKI67, TOP2A, CDK1, BRCA1, PLK1, CCNA2, CCNB1, TYMS) were found to be common across 10 hub genes, so we considered these ten genes as the key genes. The network represents highly interconnected regions of the PPIs network. The network holds 27 nodes and 350 edges
3.3 Differential expression patterns analysis of hHubGs
To investigate the differential expression patterns of hHubGs (CDK2, CHEK1, MKI67, TOP2A, CDK1, BRCA1, PLK1, CCNA2, CCNB1, TYMS) between CC and control samples, we performed box-plot analysis by using the GEPIA web-tool (see Fig. 3a). We observed the all hHubGs were significantly differentially exprerssed between CC and control samples. On the other hand, The GEPIA database was also used to investigate the prognostic power of these 10 hHubGs in CC patients using by using their survival analysis. From Fig. 3b, we observed that the lower expressions of two hHubGs (CDK2 and CCNA2) and higher expression of (CHEK1, TOP2A, BRCA1, CCNB1 and TYMS) increases the survival probability of patients, significanty.
3.4 Functional and pathway enrichment analysis of all Hub/Studied-DEGs
The GO functional enrichment analysis revealed that our proposed hHubGs significantly enriched with abundant number of biological processes (BPs), molecular functions (MFs) and cellular components (CCs) (Table 1 and Table S1). The Table 1 shows top 5 significantly enriched GO-terms for each of three categories (BPs, MFs, and CCs). These functions and pathways are highly connected with the CC related biological functional pathways in host which are crucial for developing therapeutic targets. The top five GO terms of the biological process including DNA replication, cell division, G1/S transition of mitotic cell cycle, mitotic nuclear division, and regulation of signal transduction by p53 class mediator were significantly enriched by the hHubGs-sets {BRCA1, CHEK1, CDK2, CDK1}, {CCNB2, CCNB1, CDK2, CDK1}, {TYMS, CDK2, CDK1}, {CCNB2, CDK2, CDK1}, and {BRCA1, CHEK1, CDK2}, respectively. The MFs GO terms protein binding, chromatin binding, ATP binding, protein kinase binding, and protein heterodimerization activity were significantly enriched by the hHubGs-sets {CHEK1, CCNB2, CCNB1, CDK2, CDK1}, {TOP2A, CDK1}, {TOP2A, CDK2, CDK1}, {CCNB1}, and {TOP2A}, respectively. The Cellular Components GO terms nucleoplasm, Cytosol, nucleus, spindle pole, and cytoplasm were significantly enriched by the hHubGs-sets {TOP2A, BRCA1, CHEK1, CCNB2, CCNB1, CDK2, CDK1}, {CHEK1, CCNB2, CCNB1, CDK2, CDK1}, {MKI67, CHEK1, CCNB2, CCNB1, CDK2, CDK1, TOP2A}, {CCNB1}, and {MKI67, CCNB1, CDK2, TOP2A, BRCA1}, respectively. We also observed that KEGG pathway categories Cell cycle, Pathways in cancer, HTLV-I infection, Hepatitis B, and p53 signaling pathway were significantly enriched by the hHubGs-sets {CCNB2, CCNB1, CHEK1, CDK2, CDK1}, {CDK2}, {CHEK1}, {CDK2}, and {CCNB2, CCNB1, CHEK1, CDK2, CDK1}, respectively. The other significantly enriched GO terms and KEGG pathways of hHubGs-sets were given in Table S4.
3.5 Transcriptional and post transcriptional regulatory factors of hHubGs
The network analysis of hHubGs with TFs detected top-ranked four significant TFs (TEAD1, ZBTB33, RCOR2, and ZEB1) as the key transcriptional regulatory factors for hHubGs (see Fig. 4). We found that TEAD1 is key TFs of three hHubGs (TYMS, CCNB1, and MKI67), ZBTB33 for four hHubGs (TYMS, CDK2, MKI67, and CDK1), RCOR2 for three hHubGs (TYMS, MKI67, and TOP2A), and ZEB1 for three hHubGs (BRCA1, CDK2, and MKI67). Similarly, the network analysis of hHubGs with miRNAs identified top-ranked three significant miRNAs denote as hsa-miR-548d-5p, hsa-miR-146a and hsa-miR-559 that are considered as the key post-transcriptional regulatory factors for hHubGs-sets {CCNA2, CDK2, BRCA1}, { CHEK1, TYMS, BRCA1, TOP2A}, and { CDK2, MKI67, CCNB1} respectively (see Fig. 4).
3.6 Drug repurposing by molecular docking
We considered 10 hHubGs and its regulatory 4 TFs (ZBTB33, TEAD1, ZEB1, RCOR2) as the m = 14 drug target receptors. We downloaded 3D structure of 14 receptors (BRCA1, CCNB1, CDK1, TOP2A, CCNA2, CDK2, CHEK1, MKI67, PLK1, TYMS, RCOR2, TEAD1, ZBTB33, ZEB1) from Protein Data Bank (PDB) [107] with PDB IDs 1n5o, 2b9r, 4y72, 1zxm, 1h26, 1b38, 1nvq, 1r21, 1q4o, 1hw3, 4czz, 6im5, 6df5, 2e19 respectively. The PubChem database [109] was used to retrieve the 3D structures of 77 drugs. To determine the binding scores for each pair of target proteins and meta-drug agents, molecular docking analysis was performed between m = 14 target proteins and n = 77 meta-drug agents. Then, we sorted the targets according to the row sums of the binding score matrix A = (Aij) and the drug agents according to the column sums in order to choose a small number of drug agents as the candidate drugs. The Fig. 5a showed the image of binding score matrix \({{\varvec{A}}}^{*}=\left({A}_{ij}^{*}\right)\) according to the sorted target proteins in Y-axis and n = 77 top ranked meta-drug agents in X-axis. We considered potential drug agents with a binding score of − 7.3 or less as better compounds against 14 targets. Therefore, we selected 10 top-ranked compounds (Docetaxel, Temsirolimus, Paclitaxel, Everolimus, Vincristine, Vinorelbine, cabazitaxel, Lapatinib, Irinotecan, Imatinib) as the possible candidate drug agents, possibly inhibiting the all proposed receptors for CC and these ten drugs are approved by U.S. Food and Drug Administration (FDA).
a Image of binding affinities based on the top-ordered 30 meta-drug agents out of 77 against the ordered 15 receptors, where red colors indicated the strong binding affinities, and the single star (*) indicated common receptors (published and proposed). b Image of binding affinity scores based on the ordered proposed 30 candidate-drugs in X-axis and Top-ordered 10 published proteins corresponding to CC in Y-axis
To validate our offered candidate drug agents by docking analysis with the already published targets related with CC infections, we picked up 54 papers on CC infections, those provided hub-genes and found only 10 hub-genes (ASPM, DTL, MMP1, TOP2A, PCNA, CCNB1, MCM2, AURKA, CDK1, CDC45) in which each of them was common within 4 articles out of 54 (see Table S1). We considered 10 targets corresponding to these 10 hub-genes to validate our selected drug agents against CC infections through docking analysis. From these 10 hub-genes, we observed that 3 genes (TOP2A, CDK1, CCNB1) were common with our selected hHubGs. So, the 3D structures of remain 6 hub-genes (DTL, MMP1, AURKA, PCNA, CDC45, MCM2) were downloaded from PDB with IDs 6qc0, 1cge, 1mq4, 1u76, 5dgo, 4uuz, respectively. The remain 3D structure of ASPM target was downloaded from UniProt [123] with sources ID of Q8IZT6. Then we performed docking analysis between top-ranked 30 drugs and published top-ranked 10 targets associated with CC infections. Their binding affinities (kcal/mol) were pictured in Fig. 5b. We accomplished that top-ranked 9 candidate drugs were common with our proposed top-ranked 10 candidate drugs. Finally we considered, the top-ranked 6 candidate drugs (Docetaxel, Temsirolimus, Paclitaxel, Vincristine, Everolimus, Vinorelbine) with binding affinities − 7.4 kcal/mol ≤ against the 7 published targets.
We considered top-ranked three docked complexes for protein-drug interaction profiling (see Table 2). As shown in Fig. 6a, CDK1_Docetaxel complex showed five hydrogen bonds with Leu83, Ile10, Gly11, Gln132, and Phe80 residues. Though the ligand formed major hydrophobic interactions with only two (Ala145, Leu135) residues, and three residues (Asp86, Phe80, Phe82) showed electrostatic interactions with the ligand. In Fig. 6b, CHEK1_Temsirolimus complex exposed two hydrogen bonds with Thr255, Pro250 residues, and the ligand formed electrostatic interactions with two (Glu187, Pro250) residues. In the case of the TOP2A_Paclitaxel complex, Paclitaxel showed six hydrogen bonds with Tyr82, Arg241, Arg241, Gln310, Ser320, and Gln310 residues. Paclitaxel also formed hydrophobic interactions with Ile311, Trp62, Tyr72 residues. Furthermore, Paclitaxel also formed electrostatic interactions with Glu379 residue (see Fig. 6c).
3.7 Screening drugs with proposed hHubGs for CC treatment via DGIdb
For the corss validation, Drug Gene Interaction Database (DGIdb) web resource [124] was used to retrieve drugs that interact with top-ranked 10 hHubGs. We found 9 out of the 10 genes have 456 interactions with drugs (see Fig. 7 and Supplementary Table S5). We found that, five drugs (Paclitaxel, Vincristine, Everolimus, Vinorelbine, Irinotecan) were common with our proposed top-ranked 10 drugs. BRCA1 has the highest number of inhibitory interactions among all genes with 5 drugs such as everolimus, vinorelbine, paclitaxel, and irinotecan. TOP2A can be inhibited by two drugs such as vincristine and paclitaxel. TYMS also can be inhibited by two drugs such as vincristine and irinotecan. CDK2 can be inhibited by paclitaxel.
Gene-Drug interactions based on DGIdb and the network consists of 467 nodes and 539 edges. It contains proposed top-ranked 10 hHubGs (magenta color), drugs (deep sky blue) and the five drugs in light green (Paclitaxel, Vincristine, Everolimus, Vinorelbine, Irinotecan) indicated common drugs along with our proposed top-ranked 10 drugs
3.8 Molecular dynamic simulation
Docetaxel, Temsirolimus, and Paclitaxel were the top-ranked three potential drugs out of all those suggested. Thus, these top-ranked 3 drug-agents were considered to analyze the stability through molecular dynamic simulations.
The RMSDs of three complexes and three apo form of proteins were determined from the simulation trajectories and plotted in Fig. 8a. From the figure, we observed that Paclitaxel-TOP2A obtained the highest RMSD values, while Docetaxel-CDK1 attained the lowest RMSDs in the 100 ns simulation. The figure demonstrates that docetaxel-CDK1 displayed a more stiff conformation compared to the other two proteins, reached equilibrium at 2 ns, and remained stable after that. On the other hand, the apo form of CDK1 also reached at equilibrium after 2 ns and maintained RMSD values up to 1.95 Å for 100 ns. The temsirolimus-CHEK1 complex exhibited slight fluctuations between about 40000 ps and 55000 ps and stabilized in the remaining simulations, and the apo form of CHEK1 also exhibited slight fluctuations between about 25000 ps and 50000 ps, after that it decreased to 1.4 Å and remained steady thereafter. On the contrary, the flexibility of paclitaxel-TOP2A complex increased dramatically, with RMSD values steadily increasing from 2 to 4 over time. The apo form of TOP2A reached at equilibrium after 3 ns and maintained RMSD up to 1.8 Å for 75 ns, after which the RMSD was seen to increase gradually. Here, we estimated the MM-PBSA binding energies for top-ranked 3 drugs with as mentioned previously. Figure 8b showed the binding energies for top-ranked 3 complexes (docetaxel-CDK1, temsirolimus-CHEK1, paclitaxel-TOP2A). The average binding energies of the docetaxel-CDK1, temsirolimus-CHEK1, paclitaxel-TOP2A complexes were 10.23 kJ/mol, 26.65 kJ/mol, and 7.56 kJ/mol, respectively.
a Time evolution of root-mean-square deviations (RMSDs) of backbone atoms (C, Cα, and N) for protein for top-ranked three complexes. b Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) analysis was used to determine the binding free energy (kJ/mol) of each snapshot, which represents the change in binding stability of each complex across simulations; positive values indicate better binding. Complexes: Dark blue docetaxel-CDK1, green temsirolimus-CHEK1, red paclitaxel-TOP2A, light blue apo-CDK1, light green apo-CHEK1, light red apo-TOP2A
4 Discussion
CC is the most prevalent type of malignant tumor in women where its 5-year survival rate is only about 52%. Thus, for enhancing the survival rate and minimizing the mortality rate of CC patients it is required more studies to confirm the effective biomarkers and candidate drug agents [24]. For that, the target-based therapy for cervical cancer has become a hot spot in the medication system [125]. In recent years, multiple biomarkers for cervical cancer have been discovered (see Table S2). However, the complex and varied biomarkers of targeted therapy for cervical cancer also need to be properly uncovered and investigated [126]. In this study, we collected 255 hub genes/ study genes from 52 reputed published articles to find the independent meta-receptors which closely connected with CC disease. Among them, we detected 10 hHubGs (CDK1, CDK2, CHEK1, MKI67, TOP2A, BRCA1, PLK1, CCNA2, CCNB1, TYMS) as the proposed hub/key genes with emphasis on their roles, regulatory mechanisms, and therapeutic candidates. The box-plots based on the expression profile of hHubGs through the GEPIA web server showed that hHubGs are significantly differentially expressed genes (DEGs) between CC disease and control samnples. It should be noted here that the GEPIA web server was developed based on the RNA sequencing expression profiles of tumors and normal (9,736 and 8,587)samples comprising the TCGA and the GTEx projects.
According to the existing literature, CDK2 was a more promising therapeutic target for cervical cancer [37, 59]. CHEK1 hub gene produced a high score expression value in CC tissues compared with normal tissues and was considered as a significant upregulated expressed gene (P < 0.01) [20, 44]. Yuan et al. revealed that MKI67 showed high expression in CC [44]. TOP2A is considered a potential biomarker to improve the diagnosis of CC [12, 13, 20, 24, 32, 40, 42, 44, 55, 56]. CDK1 may play a significant role in regulating the genetic network associated with cervical cancer occurrence, development, and metastasis [20, 25, 32, 40,41,42, 45, 57, 65]. PLK1 played a role in the occurrence and progression of CSCC [45]. To activate the JAK/STAT pathway BRCA1 increased the sensitivity of cervical squamous cell carcinoma (CSCC) patients to cisplatin-based CCRT with upregulated expression of STAT1 [54]. CCNB1 also enacted a key role in the development of CC under some signaling pathways [12, 25, 32, 53, 65]. Y. Liu et al. reported that the progression of CC is activated by the expression of CCNA2 [41]. Some researchers discovered that TYMS expression levels were elevated in cervical cancer and were positively connected with cervical cancer prognosis [13, 24, 57].
We considered the top five mutual GO terms including BPs, MFs and CCs, and KEGG pathways to explore the pathogenetic processes of hHubGs, those were significantly associated with cervical cancer disease based on the hub-DEGs and hHubGs. Among them the GO terms, the top five common BPs (DNA replication, cell division, G1/S transition of mitotic cell cycle, mitotic nuclear division, regulation of signal transduction by p53 class mediator) showed the significant association with CC which was supported by the previous individual studies [12, 13, 127, 128]. The top five common MFs (protein binding, chromatin binding, ATP binding, protein kinase binding, protein heterodimerization activity) were significantly associated with CC disease and that were also supported by some previous studies[12, 13, 129,130,131,132]. Similarly, top four Cellular Components (nucleoplasm, cytosol, nucleus, spindle pole, and cytoplasm) were significantly associated with CC disease which was existed by the different individual literatures [133,134,135]. We also found the top five common KEGG pathways (Cell cycle, Pathways in cancer, HTLV-I infection, Hepatitis B, and p53 signaling pathway) that were significantly enriched for CC, and it is reported by some others [12, 42, 136].
We introduced four transcriptional (TFs) and three post-transcriptional (miRNAs) regulatory factors in results sections as well as the TFs proteins (TEAD1, ZBTB33, RCOR2, and ZEB1) were used as the drug target receptors for predicting the drug agents. In literature review, the previous study suggested that TEAD1 was an significant biomarker for CC [137]. ZBTB33 was able to prevent cervical cancer cell proliferation and EMT [138]. RCOR2 was established to be functional in cancer stem cells, where it positively regulates stemness gene expression [139]. Chen J. X. et al. suggested that the effect of hypoxia-induced ZEB1-driven cancer cells induced macrophage infiltration into hypoxic area through the CCR2–NF-κB pathway showed poor connection of prognosis in CC [140].
We considered top-ranked identified proteins and their regulation 4 important TF proteins as drug target proteins and conducted molecular docking analysis with 77 meta-drug agents to find viable candidate drug agents for the medication of CC (see Table S1). Then, we choose top-ranked ten drugs (Docetaxel, Temsirolimus, Paclitaxel, Vincristine, Everolimus, Vinorelbine, cabazitaxel, Lapatinib, Irinotecan, Imatinib) as the potential drugs for CC infections based on their binding affinity scores (kCal/mol) compared with all the target receptors (see Fig. 5a). On the other hand, we also confirmed that our proposed hHubGs suggested by others. Moreover, the hHubGs matched by previous articles in favour of 10 studies for TOP2A and CDK1, 5 studies for CCNB1, 3 studies for TYMS, 2 studies for CDK2 and CHEK1, and only single study for PLK1, CCNA2, BRCA1and MKI67 genes (see Fig. 9a). The suggested drugs of our study also reported by the other researchers such as Docetaxel [70, 91], Temsirolimus [72, 74, 88, 89], Paclitaxel [14, 70, 73, 80, 82, 87, 89, 91], Vincristine [14, 70, 91], Everolimus [89], Vinorelbine [14, 73, 91], cabazitaxel [90], Lapatinib [74, 86, 88, 89], Irinotecan [73, 91], Imatinib[88,89,90] for the treatment of CC infections (see Fig. 9b). On the other hand, we used DGIdb online database to screen the drugs that interact with our proposed 10 hHubGs for cross validation and found that five drugs (Paclitaxel, Vincristine, Everolimus, Vinorelbine, Irinotecan) were common with our proposed top-ranked 10 drugs. Then, we also validated top-ranked 30 candidate-drugs against the top-ranked 10 published receptors (ASPM, DTL, MMP1, AURKA, PCNA, CCNB1, CDC45, MCM2, TOP2A, CDK1) associated with CC infections by molecular docking analysis and found their strongly significant binding affinity scores with top-ranked 6 candidate-drugs (see Fig. 5b). Finally, we investigated the stability of top-ranked three drugs (Docetaxel, Temsirolimus, Paclitaxel) by using 100 ns MD-based MM-PBSA simulations for three top-ranked proposed receptors (CDK1, CHEK1, TOP2A), and observed their stable performance according to the laws of physics [141, 142]. Therefore, the proposed candidate drugs might be played a vital role in the treatment of CC infections.
5 Conclusions
The current work used a variety of well-known bioinformatics tools to discover hub of the HubGs (hHubGs) highlighting their regulatory factors and dysregulated molecular functions and pathways pathways that are responsible for CC development. At first, we collected 255 HubGs/studied-genes that were published by the individual studies.. Among them, we selected 10 HubGs (CDK1, CDK2, CHEK1, MKI67, TOP2A, BRCA1, PLK1, CCNA2, CCNB1, TYMS) as the hHubGs by the PPI network analysis and validated their differential expression patterns between CC and normal samples through the GPEA database. The gene ontology (GO) and KEGG pathway enrichment analysis of HubGs revealed some crucial CC-causing BPs (DNA replication, cell division), MFs (protein binding, chromatin binding) and CCs (nucleoplasm, cytosol) by involving hHubGs. The gene regulatory network (GRN) analysis exposed four TFs proteins (TEAD1, ZBTB33, RCOR2, and ZEB1) and three miRNAs (hsa-miR-548d-5p, hsa-miR-146a and hsa-miR-559) as the key transcriptional and post-transcriptional regulators of hHubGs. Then, we identified hHubGs-guided top-ranked FDA-approved 10 candidate drugs (Docetaxel, Temsirolimus, Paclitaxel, Vincristine, Everolimus, Vinorelbine, cabazitaxel, Lapatinib, Irinotecan, Imatinib) and validated them against the state-of-the-arts independent receptors by molecular docking analysis. Finally, we offered possible candidate drug agents, such as Docetaxel, Temsirolimus, Paclitaxel, and examined their stability performance by using 100 ns MD-based MM-PBSA simulations for the top-ranked three proposed proteins (CDK1, CHEK1, TOP2A), and also detected their stable performance. Hence, the selected genetic biomarkers and candidate repurposing drugs derived from this study has merit for CC disease diagnosis and therapies.
Availability of data and materials
All data analyzed during this study are included in this article and its additional files.
Abbreviations
- CC:
-
Cervical cancer
- CSCC:
-
Cervical squamous cell carcinoma
- HPV:
-
Human Papillomavirus
- PPI:
-
Protein–Protein Interaction
- ENCODE:
-
Encyclopedia Of DNA Elements
- MCODE:
-
Molecular Complex Detection
- DEGs:
-
Differentially Expressed Genes
- cDEGs:
-
Common Differentially Expressed Genes
- cHubGs:
-
Common Hub Genes
- cHubPs:
-
Common Hub Proteins
- hHubGs:
-
Hub of the hub-genes
- KPs:
-
Key proteins
- GO:
-
Gene ontology
- BPs:
-
Biological processes
- MFs:
-
Molecular function
- CCs:
-
Cellular components
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- TFs:
-
Transcription factors
- miRNAs:
-
Micro-RNAs
- MD:
-
Molecular dynamic
- MM-PBSA:
-
Molecular Mechanics Poisson–Boltzmann Surface Area
- RMSD:
-
Root mean square deviation
- 3D:
-
Three-Dimensional
- PDB:
-
Protein data bank
- FDA:
-
U.S. Food and Drug Administration
- YASARA:
-
Yet Another Scientific Artificial Reality Application
References
Mattiuzzi C, Lippi G. Current cancer epidemiology. J Epidemiol Glob Health. 2019. https://doi.org/10.2991/jegh.k.191008.001.
Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9:665.
Lopez MS, Baker ES, Maza M, Fontes-Cintra G, Lopez A, Carvajal JM, Nozar F, Fiol V, Schmeler KM. Cervical cancer prevention and treatment in Latin America. J Surg Oncol. 2017;115:615.
Shi X, Wang J, Dai S, Qin L, Zhou J, Chen Y. Apolipoprotein C1 (Apoc1): a novel diagnostic and prognostic biomarker for cervical cancer. Onco Targets Ther. 2020. https://doi.org/10.2147/OTT.S280690.
Goodman A. HPV testing as a screen for cervical cancer. BMJ. 2015;350:h2372.
Wardak S. Human Papillomavirus (HPV) and cervical cancer. Med Dosw Mikrobiol. 2016. https://doi.org/10.12968/indn.2020.2.20.
Chávez-lópez MG, Zúñiga-garcía V, Castro-magdonel BE, Vera E, Garrido E, Sánchez-ramos J, Ponce-castañeda MV, Cabrera-muñoz ML, Escobar Y, Ortiz CS, et al. Eag1 gene and protein expression in human retinoblastoma tumors and its regulation by PRb in HeLa cells. Genes (Basel). 2020. https://doi.org/10.3390/genes11020119.
Somashekhar SP, Ashwin KR. Management of early stage cervical cancer. Rev Recent Clin Trials. 2015. https://doi.org/10.2174/1574887110666150923113629.
Brucker SY, Ulrich UA. Surgical treatment of early-stage cervical cancer. Oncol Res Treat. 2016;39:508.
Falcetta FS, Medeiros LR, Edelweiss MI, Pohlmann PR, Stein AT, Rosa DD. Adjuvant platinum-based chemotherapy for early stage cervical cancer. Cochrane Database Syst Rev. 2016. https://doi.org/10.1002/14651858.CD005342.pub4.
Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JWW, Comber H, Forman D. Cancer incidence and mortality patterns in europe: estimates for 40 countries in 2012. Eur J Cancer. 2013. https://doi.org/10.1016/j.ejca.2012.12.027.
Wu X, Peng L, Zhang Y, Chen S, Lei Q, Li G, Zhang C. Identification of key genes and pathways in cervical cancer by bioinformatics analysis. Int J Med Sci. 2019. https://doi.org/10.7150/ijms.34172.
Yang H, Xue J, Li J, Wan L, Zhu Y. identification of key genes and pathways of diagnosis and prognosis in cervical cancer by bioinformatics analysis. Mol Genet Genomic Med. 2020. https://doi.org/10.1002/mgg3.1200.
Reza MS, Harun-Or-Roshid M, Islam MA, Hossen MA, Hossain MT, Feng S, Xi W, Mollah MNH, Wei Y. Bioinformatics screening of potential biomarkers from MRNA expression profiles to discover drug targets and agents for cervical cancer. Int J Mol Sci. 2022;23:3968. https://doi.org/10.3390/ijms23073968.
Mosharaf MP, Reza MS, Kibria MK, Ahmed FF, Kabir MH, Hasan S, Mollah MNH. Computational identification of host genomic biomarkers highlighting their functions, pathways and regulators that influence SARS-CoV-2 infections and drug repurposing. Sci Rep. 2022. https://doi.org/10.1038/s41598-022-08073-8.
Chang JW, Ding Y, Qamar MT, Shen Y, Gao J, Chen LL. A Deep learning model based on sparse auto-encoder for prioritizing cancer-related genes and drug target combinations. Carcinogenesis. 2019. https://doi.org/10.1093/carcin/bgz044.
Ahmed FF, Reza MS, Sarker MS, Islam MS, Mosharaf MP, Hasan S, Mollah MNH. Identification of Host Transcriptome-Guided Repurposable Drugs for SARS-CoV-1 Infections and Their Validation with SARS-CoV-2 infections by using the integrated bioinformatics approaches. PLoS ONE. 2022;17: e0266124. https://doi.org/10.1371/journal.pone.0266124.
Reza MS, Zhang H, Hossain MT, Jin L, Feng S, Wei Y. Comtop: protein residue-residue contact prediction through mixed integer linear optimization. Membranes (Basel). 2021. https://doi.org/10.3390/membranes11070503.
Mao D, Xu R, Chen H, Chen X, Li D, Song S, He Y, Wei Z, Zhang C. Cross-talk of focal adhesion-related gene defines prognosis and the immune microenvironment in gastric cancer. Front Cell Dev Biol. 2021. https://doi.org/10.3389/fcell.2021.716461.
Wu B, Xi S. Bioinformatics analysis of differentially expressed genes and pathways in the development of cervical cancer. BMC Cancer. 2021. https://doi.org/10.1186/s12885-021-08412-4.
Moutal A, Martin LF, Boinon L, Gomez K, Ran D, Zhou Y, Stratton HJ, Cai S, Luo S, Gonzalez KB, et al. SARS-CoV-2 Spike Protein Co-Opts VEGF-A/Neuropilin-1 receptor signaling to induce analgesia. Pain. 2021;162:243–52. https://doi.org/10.1097/j.pain.0000000000002097.
He Z, Wang X, Yang Z, Jiang Y, Li L, Wang X, Song Z, Wang X, Wan J, Jiang S, et al. Expression and Prognosis of CDC45 in cervical cancer based on the GEO database. PeerJ. 2021. https://doi.org/10.7717/peerj.12114.
Mei Y, Jiang P, Shen N, Fu S, Zhang J. Identification of MiRNA-MRNA regulatory network and construction of prognostic signature in cervical cancer. DNA Cell Biol. 2020. https://doi.org/10.1089/dna.2020.5452.
Qiu HZ, Huang J, Xiang CC, Li R, Zuo ED, Zhang Y, Shan L, Cheng X. Screening and discovery of new potential biomarkers and small molecule drugs for cervical cancer: a bioinformatics analysis. Technol Cancer Res Treat. 2020. https://doi.org/10.1177/1533033820980112.
Yi Y, Fang Y, Wu K, Liu Y, Zhang W. Comprehensive gene and pathway analysis of cervical cancer progression. Oncol Lett. 2020. https://doi.org/10.3892/ol.2020.11439.
Deng SP, Zhu L, Huang DS. Predicting Hub genes associated with cervical cancer through gene co-expression networks. IEEE/ACM Trans Comput Biol Bioinforma. 2016. https://doi.org/10.1109/TCBB.2015.2476790.
Wang J, Zheng H, Han Y, Wang G, Li Y. A novel four-gene prognostic signature as a risk biomarker in cervical cancer. Int J Genomics. 2020. https://doi.org/10.1155/2020/4535820.
Liu J, Yang J, Gao F, Li S, Nie S, Meng H, Sun R, Wan Y, Jiang Y, Ma X, et al. A MicroRNA-Messenger RNA regulatory network and its prognostic value in cervical cancer. DNA Cell Biol. 2020. https://doi.org/10.1089/dna.2020.5590.
Ouyang D, Ouyang D, Yang P, Cai J, Sun S, Wang Z. Comprehensive analysis of prognostic alternative splicing signature in cervical cancer. Cancer Cell Int. 2020. https://doi.org/10.1186/s12935-020-01299-4.
Chen H, Wang X, Jia H, Tao Y, Zhou H, Wang M, Wang X, Fang X. Bioinformatics analysis of key genes and pathways of cervical cancer. Onco Targets Ther. 2020;13:13275–83. https://doi.org/10.2147/OTT.S281533.
Xue H, Sun Z, Wu W, Du D, Liao S. Identification of Hub Genes as potential prognostic biomarkers in cervical cancer using comprehensive bioinformatics analysis and validation studies. Cancer Manag Res. 2021. https://doi.org/10.2147/CMAR.S282989.
Zhao Q, Li H, Zhu L, Hu S, Xi X, Liu Y, Liu J, Zhong T. Bioinformatics Analysis Shows That Top2a functions as a key candidate gene in the progression of cervical cancer. Biomed Reports. 2020. https://doi.org/10.3892/br.2020.1328.
Ma X, Liu J, Wang H, Jiang Y, Wan Y, Xia Y, Cheng W. Identification of crucial aberrantly methylated and differentially expressed genes related to cervical cancer using an integrated bioinformatics analysis. 2020. Biosci Rep. https://doi.org/10.1042/BSR20194365.
Mallik S, Seth S, Bhadra T, Zhao Z. A Linear regression and deep learning approach for detecting reliable genetic alterations in cancer using dna methylation and gene expression data. Genes (Basel). 2020. https://doi.org/10.3390/genes11080931.
Liu J, Li S, Lin L, Jiang Y, Wan Y, Zhou S, Cheng W. Co-Expression Network Analysis Identified Atypical Chemokine Receptor 1 (ACKR1) association with lymph node metastasis and prognosis in cervical cancer. Cancer Biomarkers. 2020. https://doi.org/10.3233/CBM-190533.
Tu S, Zhang H, Yang X, Wen W, Song K, Yu X, Qu X. Screening of cervical cancer-related hub genes based on comprehensive bioinformatics analysis. Cancer Biomark. 2021. https://doi.org/10.3233/cbm-203262.
Wu K, Yi Y, Liu F, Wu W, Chen Y, Zhang W. Identification of key pathways and genes in the progression of cervical cancer using bioinformatics analysis. Oncol Lett. 2018. https://doi.org/10.3892/ol.2018.8768.
Liu J, Wu Z, Wang Y, Nie S, Sun R, Yang J, Cheng W. A prognostic signature based on immune-related genes for cervical squamous cell carcinoma and endocervical adenocarcinoma. Int Immunopharmacol. 2020. https://doi.org/10.1016/j.intimp.2020.106884.
Liu J, Liu S, Yang X. Construction of Gene modules and analysis of prognostic biomarkers for cervical cancer by weighted gene co-expression network analysis. Front Oncol. 2021. https://doi.org/10.3389/fonc.2021.542063.
Xu Z, Zhou Y, Shi F, Cao Y, Dinh TLA, Wan J, Zhao M. Investigation of differentially-expressed MicroRNAs and genes in cervical cancer using an integrated bioinformatics analysis. Oncol Lett. 2017. https://doi.org/10.3892/ol.2017.5766.
Liu Y, Yi Y, Wu W, Wu K, Zhang W. Bioinformatics prediction and analysis of hub genes and pathways of three types of gynecological cancer. Oncol Lett. 2019. https://doi.org/10.3892/ol.2019.10371.
Xue JM, Liu Y, Wan LH, Zhu YX. Comprehensive analysis of differential gene expression to identify common gene signatures in multiple cancers. Med Sci Monit. 2020. https://doi.org/10.12659/MSM.919953.
Wang M, Li L, Liu J, Wang J. A gene interaction network-based method to measure the common and heterogeneous mechanisms of gynecological cancer. Mol Med Rep. 2018. https://doi.org/10.3892/mmr.2018.8961.
Yuan Y, Shi X, Li B, Peng M, Zhu T, Lv G, Liu L, Jin H, Li L, Qin D. Integrated Analysis of Key MicroRNAs /TFs /MRNAs/ in HPV-positive cervical cancer based on microrna sequencing and bioinformatics analysis. Pathol Res Pract. 2020. https://doi.org/10.1016/j.prp.2020.152952.
Mousavi SZ, Poortahmasebi V, Mokhtari-Azad T, Shahmahmoodi S, Farahmand M, Farzanehpour M, Jalilvand S. The dysregulation of microarray gene expression in cervical cancer is associated with overexpression of a unique messenger RNA signature. Iran J Microbiol. 2020. https://doi.org/10.18502/ijm.v12i6.5039.
Zhang X, Wang Y. Identification of hub genes and key pathways associated with the progression of gynecological cancer. Oncol Lett. 2019. https://doi.org/10.3892/ol.2019.11004.
Chen Q, Zeng X, Huang D, Qiu X. Identification of differentially expressed MiRNAs in early-stage cervical cancer with lymph node metastasis across the cancer genome atlas datasets. Cancer Manag Res. 2018. https://doi.org/10.2147/CMAR.S183488.
Li S, Han F, Qi N, Wen L, Li J, Feng C, Wang Q. Determination of a six-gene prognostic model for cervical cancer based on WGCNA combined with LASSO and Cox-PH Analysis. World J Surg Oncol. 2021;19:1–11. https://doi.org/10.1186/s12957-021-02384-2.
Fu XH, Wu YF, Xue F. Probing pathway-related modules in invasive squamous cervical cancer based on topological centrality of network strategy. J Cancer Res Ther. 2018. https://doi.org/10.4103/0973-1482.187352.
Wu B, Xi S. Bioinformatics analysis of the transcriptional expression of minichromosome maintenance proteins as potential indicators of survival in patients with cervical cancer. BMC Cancer. 2021. https://doi.org/10.1186/s12885-021-08674-y.
Meng H, Liu J, Qiu J, Nie S, Jiang Y, Wan Y, Cheng W. Identification of key genes in association with progression and prognosis in cervical squamous cell carcinoma. DNA Cell Biol. 2020. https://doi.org/10.1089/dna.2019.5202.
Ding H, Zhang L, Zhang C, Song J, Jiang Y. Screening of significant biomarkers related to prognosis of cervical cancer and functional study based on LncRNA-Associated CeRNA Regulatory Network. Comb Chem High Throughput Screen. 2020. https://doi.org/10.2174/1386207323999200729113028.
Li S, Liu N, Piao J, Meng F, Li Y. Ccnb1 expedites the progression of cervical squamous cell carcinoma via the regulation by Foxm1. Onco Targets Ther. 2020. https://doi.org/10.2147/OTT.S279951.
Wen X, Liu S, Cui M. Effect of BRCA1 on the concurrent chemoradiotherapy resistance of cervical squamous cell carcinoma based on transcriptome sequencing analysis. Biomed Res Int. 2020. https://doi.org/10.1155/2020/3598417.
Suman S, Mishra A. Network analysis revealed aurora kinase dysregulation in five gynecological types of cancer. Oncol Lett. 2018. https://doi.org/10.3892/ol.2017.7368.
Zhang X, Bai J, Yuan C, Long L, Zheng Z, Wang Q, Chen F, Zhou Y. Bioinformatics analysis and identification of potential genes related to pathogenesis of cervical intraepithelial neoplasia. J Cancer. 2020. https://doi.org/10.7150/jca.38211.
Sun D, Han L, Cao R, Wang H, Jiang J, Deng Y, Yu X. Prediction of a MiRNA-MRNA functional synergistic network for cervical squamous cell carcinoma. FEBS Open Bio. 2019. https://doi.org/10.1002/2211-5463.12747.
Oany AR, Mia M, Pervin T, Alyami SA, Moni MA. Integrative systems biology approaches to identify potential biomarkers and pathways of cervical cancer. J Pers Med. 2021. https://doi.org/10.3390/jpm11050363.
Xiao L, Zhang S, Zheng Q, Zhang S. Dysregulation of KIF14 regulates the cell cycle and predicts poor prognosis in cervical cancer: a study based on integrated approaches. Brazilian J Med Biol Res. 2021;54:1–10. https://doi.org/10.1590/1414-431X2021e11363.
Yu D, Li Y, Ming Z, Wang H, Dong Z, Qiu L, Wang T. Comprehensive circular RNA expression profile in radiation-treated hela cells and analysis of radioresistance-related circRNAs. PeerJ. 2018. https://doi.org/10.7717/peerj.5011.
Zhang X, Yang P, Luo X, Su C, Chen Y, Zhao L, Wei L, Zeng H, Varghese Z, Moorhead JF, et al. High olive oil diets enhance cervical tumour growth in mice: transcriptome analysis for potential candidate genes and pathways. Lipids Health Dis. 2019. https://doi.org/10.1186/s12944-019-1023-6.
Xu F, Shen J, Xu S. Multi-Omics data analyses construct a six immune-related genes prognostic model for cervical cancer in tumor microenvironment. Front Genet. 2021. https://doi.org/10.3389/fgene.2021.663617.
Jiang P, Cao Y, Gao F, Sun W, Liu J, Ma Z, Xie M, Fu S. SNX10 and PTGDS are associated with the progression and prognosis of cervical squamous cell carcinoma. BMC Cancer. 2021. https://doi.org/10.1186/s12885-021-08212-w.
Yang C, Xu X, Jin H. Identification of Potential MiRNAs and candidate genes of cervical intraepithelial neoplasia by bioinformatic analysis. Eur J Gynaecol Oncol. 2016. https://doi.org/10.12892/ejgo3131.2016.
Luo H, Li Y, Zhao Y, Chang J, Zhang X, Zou B, Gao L, Wang W. Comprehensive analysis of CircRNA expression profiles during cervical carcinogenesis. Front Oncol. 2021;11:1–13. https://doi.org/10.3389/fonc.2021.676609.
Tong Y, Sun P, Yong J, Zhang H, Huang Y, Guo Y, Yu J, Zhou S, Wang Y, Wang Y, et al. Radiogenomic analysis of papillary thyroid carcinoma for prediction of cervical lymph node metastasis: a preliminary study. Front Oncol. 2021. https://doi.org/10.3389/fonc.2021.682998.
Zhang Z, Zhao S, Wang K, Shang M, Chen Z, Yang H, Chen Y, Chen B. Identification of biomarkers associated with cervical lymph node metastasis in papillary thyroid carcinoma: evidence from an integrated bioinformatic analysis. Clin Hemorheol Microcirc. 2021. https://doi.org/10.3233/CH-201074.
Venkataramnan S, Izam WN. Cervical cancer and gene expression analysis with key genes identification by computational method. J Bio Innov. 2020. https://doi.org/10.46344/jbino.2020.v09i05.26.
Liu J, Nie S, Gao M, Jiang Y, Wan Y, Ma X, Zhou S, Cheng W. Identification of EPHX2 and RMI2 as two novel key genes in cervical squamous cell carcinoma by an integrated bioinformatic analysis. J Cell Physiol. 2019. https://doi.org/10.1002/jcp.28731.
Meneur C, Eswaran S, Adiga D, Sriharikrishnaa S, Nadeem K, Mallya S, Chakrabarty S, Kabekkodu P. Analysis of nuclear encoded mitochondrial gene networks in cervical cancer. Asian Pacific J Cancer Prev. 2021. https://doi.org/10.1002/jcp.28731.
Mei J, Xing Y, Lv J, Gu D, Pan J, Zhang Y, Liu J. Construction of an immune-related gene signature for prediction of prognosis in patients with cervical cancer. Int Immunopharmacol. 2020. https://doi.org/10.1016/j.intimp.2020.106882.
Klopp AH, Eifel PJ. Chemoradiotherapy for Cervical Cancer in 2010. Curr Oncol Rep. 2011. https://doi.org/10.1007/s11912-010-0134-z.
Tao X, Hu W, Ramirez PT, Kavanagh JJ. Chemotherapy for recurrent and metastatic cervical cancer. Gynecol Oncol. 2008. https://doi.org/10.1016/j.ygyno.2008.04.024.
Diaz-Padilla I, Monk BJ, Mackay HJ, Oaknin A. Treatment of metastatic cervical cancer: future directions involving targeted agents. Crit Rev Oncol Hematol. 2013;85:303.
Tierney JF, Vale C, Symonds P. Concomitant and neoadjuvant chemotherapy for cervical cancer. Clin Oncol. 2008. https://doi.org/10.1016/j.clon.2008.04.003.
Verschraegen CF. Irinotecan for the treatment of cervical cancer. Oncology (Williston Park). 2002;16:32.
Su J, Zhang F, Li X, Liu Z. Osthole Promotes the Suppressive Effects of Cisplatin on NRF2 expression to prevent drug-resistant cervical cancer progression. Biochem Biophys Res Commun. 2019. https://doi.org/10.1016/j.bbrc.2019.04.021.
Ackermann S, Beckmann MW, Thiel F, Bogenrieder T. Topotecan in cervical cancer. Int J Gynecol Cancer. 2007;17:1215.
Chandimali NA, Sun HN, Park YH, Kwon T. Suppresses cervical cancer stem cell characteristics and progression by inhibiting. In Vivo (Brooklyn). 2020. https://doi.org/10.21873/invivo.11879.
Markman M. Advances in cervical cancer pharmacotherapies. Expert Rev Clin Pharmacol. 2014;7:219.
Moga MA, Dima L, Balan A, Blidaru A, Dimienescu OG, Podasca C, Toma S. Are bioactive molecules from seaweeds a novel and challenging option for the prevention of HPV infection and cervical cancer therapy?—a review. Int J Mol Sci. 2021;22:629.
Lee SW, Kim YM, Kim MB, Kim DY, Kim JH, Nam JH, Kim YT. Chemosensitivity of uterine cervical cancer demonstrated by the histoculture drug response assay. Tohoku J Exp Med. 2009. https://doi.org/10.1620/tjem.219.277.
Small W. Potential for use of amifostine in cervical cancer. Semin Oncol. 2002. https://doi.org/10.1053/sonc.2002.37366.
Ai Z, Wang J, Xu Y, Teng Y. Bioinformatics analysis reveals potential candidate drugs for cervical cancer. J Obstet Gynaecol Res. 2013. https://doi.org/10.1111/jog.12022.
Ujhelyi Z, Kalantari A, Vecsernyés M, Róka E, Fenyvesi F, Póka R, Kozma B, Bácskay I. The enhanced inhibitory effect of different antitumor agents in self-microemulsifying drug delivery systems on human cervical cancer HeLa Cells. Molecules. 2015. https://doi.org/10.3390/molecules200713226.
Marquina G, Manzano A, Casado A. Targeted agents in cervical cancer: beyond Bevacizumab. Curr Oncol Rep. 2018. https://doi.org/10.1007/s11912-018-0680-3.
Sharma S, Deep A, Sharma AK. Current treatment for cervical cancer: an update. Anticancer Agents Med Chem. 2020. https://doi.org/10.2174/1871520620666200224093301.
Duenas-Gonzalez A, Gonzalez-Fierro A. Pharmacodynamics of current and emerging treatments for cervical cancer. Expert Opin Drug Metab Toxicol. 2019. https://doi.org/10.1080/17425255.2019.1648431.
Barra F, Lorusso D, Leone UM, Ditto A, Bogani G, Raspagliesi F, Ferrero S. investigational drugs for the treatment of cervical cancer. Expert Opin Investig Drugs. 2017;26:389.
Serrano-Olvera A, Cetina L, Coronel J, Dueñas-González A. Emerging drugs for the treatment of cervical cancer. Expert Opin Emerg Drugs. 2015;20:165.
Kamura T, Ushijima K. Chemotherapy for advanced or recurrent cervical cancer. Taiwan J Obstet Gynecol. 2013. https://doi.org/10.1016/j.tjog.2013.04.003.
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, et al. The STRING Database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;2011:39. https://doi.org/10.1093/nar/gkq973.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003. https://doi.org/10.1101/gr.1239303.
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014. https://doi.org/10.1186/1752-0509-8-S4-S11.
Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001. https://doi.org/10.1038/35075138.
Pržulj N, Wigle DA, Jurisica I. Functional topology in a network of protein interactions. Bioinformatics. 2004. https://doi.org/10.1093/bioinformatics/btg415.
Freeman LC. A set of measures of centrality based on betweenness. Sociometry. 1977. https://doi.org/10.2307/3033543.
Shimbel A. Structural parameters of communication networks. Bull Math Biophys. 1953. https://doi.org/10.1007/BF02476438.
Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003. https://doi.org/10.1186/1471-2105-4-2.
Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::Termfinder-Open Source Software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes introduction: motivation and design. Bioinforma Appl Note. 2004;20:3710–5. https://doi.org/10.1093/bioinformatics/bth456.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27.
Doms A, Schroeder M. GoPubMed: exploring PubMed with the gene ontology. Nucleic Acids Res. 2005. https://doi.org/10.1093/nar/gki470.
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003. https://doi.org/10.1186/gb-2003-4-9-r60.
Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019. https://doi.org/10.1093/nar/gkz240.
Feingold EA, Good PJ, Guyer MS, Kamholz S, Liefer L, Wetterstrand K, Collins FS, Gingeras TR, Kampa D, Sekinger EA, et al. The ENCODE (ENCyclopedia of DNA Elements) Project. Science. 2004;306:636.
Liu ZP, Wu C, Miao H, Wu H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database. 2015. https://doi.org/10.1093/database/bav095.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235.
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, De Beer TAP, Rempfer C, Bordoli L, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018. https://doi.org/10.1093/nar/gky427.
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, PubChem YuB, et al. Update: improved access to chemical data. Nucleic Acids Res. 2019. https://doi.org/10.1093/nar/gky1033.
Visualizer, D.S. v4. 0. 100. 13345 Accelrys Sof Tware Inc (2005).
Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007. https://doi.org/10.1093/nar/gkm276.
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: A Server for Estimating PKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005. https://doi.org/10.1093/nar/gki464.
Morris GM, Huey R, Lindstrom W, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785.
Oleg T, Arthur J, O. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010. https://doi.org/10.1002/jcc.21334.
Delano WL, Bromberg S. PyMOL User’s Guide. Californias: DeLano Scientific LLC; 2004.
Krieger, Elmar, G.V.; Spronk, C. YASARA - Yet Another Scientific Artificial Reality Application. YASARA.org 2013.
Dickson CJ, Madej BD, Skjevik ÅA, Betz RM, Teigen K, Gould IR, Walker RC. Lipid14: the amber lipid force field. J Chem Theory Comput. 2014. https://doi.org/10.1021/ct4010307.
Stewart JJP. MOPAC: a semiempirical molecular orbital program. J Comput Aided Mol Des. 1990. https://doi.org/10.1007/BF00128336.
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983. https://doi.org/10.1063/1.445869.
Krieger E, Koraimann G, Vriend G. Increasing the Precision of Comparative Models with YASARA NOVA—a self-parameterizing force field. Proteins Struct Funct Genet. 2002. https://doi.org/10.1002/prot.10104.
Mitra S, Dash R. Structural dynamics and quantum mechanical aspects of shikonin derivatives as CREBBP bromodomain inhibitors. J Mol Graph Model. 2018. https://doi.org/10.1016/j.jmgm.2018.04.014.
Srinivasan E, Rajasekaran R. Computational investigation of curcumin, a natural polyphenol that inhibits the destabilization and the aggregation of human SOD1 Mutant (Ala4Val). RSC Adv. 2016. https://doi.org/10.1039/c6ra21927f.
The Uniprot Consortium. UniProt: A worldwide hub of protein knowledge The UniProt Consortium. Nucleic Acids Res. 2019. https://doi.org/10.1093/nar/gky1049.
Freshour SL, Kiwala S, Cotto KC, Coffman AC, McMichael JF, Song JJ, Griffith M, Griffith OL, Wagner AH. Integration of the Drug-Gene Interaction Database (DGIdb 40) with open crowdsource efforts. Nucleic Acids Res. 2021;49:D1144–51. https://doi.org/10.1093/nar/gkaa1084.
Kumar L, Harish P, Malik PS, Khurana S. Chemotherapy and targeted therapy in the management of cervical cancer. Curr Probl Cancer. 2018;42:120.
Kori M, Arga KY. Potential biomarkers and therapeutic targets in cervical cancer: insights from the meta-analysis of transcriptomics data within network biomedicine perspective. PLoS One. 2018. https://doi.org/10.1371/journal.pone.0200717.
Chen H, Zhang Q, Qiao L, Fan X, Zhang W, Zhao W, Chen JJ. Cdc6 contributes to abrogating the G1 Checkpoint under hypoxic conditions in HPV E7 expressing cells. Sci Rep. 2017. https://doi.org/10.1038/s41598-017-03060-w.
Shao XY, Dong J, Zhang H, Wu YS, Zheng L. Prognostic value and potential role of alternative MRNA splicing events in cervical cancer. Front Genet. 2020. https://doi.org/10.3389/fgene.2020.00726.
Chen Z, Ling K, Zhu Y, Deng L, Li Y, Liang Z. Rucaparib antagonize multidrug resistance in cervical cancer cells through blocking the function of ABC transporters. Gene. 2020. https://doi.org/10.1016/j.gene.2020.145000.
Murugesan M, Premkumar K. Integrative MiRNA-MRNA functional analysis identifies MiR-182 as a potential prognostic biomarker in breast cancer. Mol Omi. 2021. https://doi.org/10.1039/d0mo00160k.
Xing Z, Luo Z, Yang H, Huang Z, Liang X. Screening and identification of key biomarkers in adrenocortical carcinoma based on bioinformatics analysis. Oncol Lett. 2019. https://doi.org/10.3892/ol.2019.10817.
Xie Y, Mu C, Kazybay B, Sun Q, Kutzhanova A, Nazarbek G, Xu N, Nurtay L, Wang Q, Amin A, et al. Network pharmacology and experimental investigation of Rhizoma Polygonati Extract targeted kinase with herbzyme activity for potent drug delivery. Drug Deliv. 2021. https://doi.org/10.1080/10717544.2021.1977422.
Gao C, Zhou C, Zhuang J, Liu L, Liu C, Li H, Liu G, Wei J, Sun C. MicroRNA expression in cervical cancer: novel diagnostic and prognostic biomarkers. J Cell Biochem. 2018. https://doi.org/10.1002/jcb.27029.
Ostrowska KM, Garcia A, Meade AD, Malkin A, Okewumi I, O’Leary JJ, Martin C, Byrne HJ, Lyng FM. Correlation of P16INK4A Expression and HPV copy number with cellular FTIR spectroscopic signatures of cervical cancer cells. Analyst. 2011. https://doi.org/10.1039/c0an00910e.
Von Knebel Doeberitz M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer. 2002;38:2229–42. https://doi.org/10.1016/S0959-8049(02)00462-8.
Liang Q, Yu Q, Wu H, Zhu YZ, Zhang AH. Metabolite fingerprint analysis of cervical cancer using LC-QTOF/MS and multivariate data analysis. Anal Methods. 2014. https://doi.org/10.1039/c4ay00399c.
Yamaguchi N. Multiple roles of vestigial-like family members in tumor development. Front Oncol. 2020. https://doi.org/10.3389/fonc.2020.01266.
Feng J. Upregulation of MicroRNA-4262 targets KaiSO (ZBTB33) to inhibit the proliferation and EMT of cervical cancer cells. Oncol Res. 2018. https://doi.org/10.3727/096504017X15021536183526.
Karakaidos P, Verigos J, Magklara A. Lsd1/Kdm1a, a gate-keeper of cancer stemness and a promising therapeutic target. Cancers (Basel). 2019. https://doi.org/10.3390/cancers11121821.
Chen XJ, Deng YR, Wang ZC, Wei WF, Zhou CF, Zhang YM, Yan RM, Liang LJ, Zhong M, Liang L, et al. Hypoxia-Induced ZEB1 promotes cervical cancer progression via CCL8-dependent tumour-associated macrophage recruitment. Cell Death Dis. 2019. https://doi.org/10.1038/s41419-019-1748-1.
Blatt JM, Weisskopf VF, Critchfield CL. Theoretical nuclear physics. Am J Phys. 1953. https://doi.org/10.1119/1.1933407.
Lovering AL, Seung SL, Kim YW, Withers SG, Strynadka NCJ. Mechanistic and structural analysis of a family 31 α-glycosidase and its glycosyl-enzyme intermediate. J Biol Chem. 2005. https://doi.org/10.1074/jbc.M410468200.
Acknowledgements
Not applicable.
Funding
This works was partly supported by Bangladesh Medical Research Council (BMRC) research project (Ref: BMRC/HPNSP-Research Grant/2020–2021/306(1–28)), Govt. of Bangladesh.
Author information
Authors and Affiliations
Contributions
Conceptualization, MSR and MNHM; methodology MSR; software, MSR; validation, MSR, and MNHM; formal analysis, MSR; investigation, MSR, and MNHM; data curation, MSR and MHOR; writing—original draft preparation, MSR; writing—review and editing, MSR, MHK, MHOR, MAH, MAS, and MNHM; visualization, MSR; supervision, MNHM; project administration, MNHM; funding acquisition, MNHM; All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
. Different hub/studied genes list for CC infection published by different papers in different international reputed journals. Table S2. 77 meta-drug agents for the treatment against CC. Table S4. The top 20 significantly (p-value<0.001) enriched GO functions and KEGG pathways by cDEGs involving KGs with CC diseases. Table S5. Gene-Drug interactions analysis based on our proposed hHubGs using DGIdb database.
Additional file 2: Table S3
. The MCODE raw file was selected from the PPI network through Cytoscape software, where the degree cutoff was used 80.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Reza, M.S., Hossen, M.A., Harun-Or-Roshid, M. et al. Metadata analysis to explore hub of the hub-genes highlighting their functions, pathways and regulators for cervical cancer diagnosis and therapies. Discov Onc 13, 79 (2022). https://doi.org/10.1007/s12672-022-00546-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-022-00546-6