Abstract
Background
Takeda G protein receptor 5 (TGR5) is widely recognized as a potential drug target for the treatment of metabolic diseases. TGR5 is not only a metabolic regulator, but also has a potential role that participating in developing and progressing of gastrointestinal cancer. We aimed to investigate the potential role of TGR5 in pancreatic cancer by utilizing molecular experiments and the liquid chromatography mass spectrometry (LC–MS) based metabolomics.
Methods
Herein, we assessed pancreatic cancer proliferation, migration and invasion in response to TGR5 antagonist SBI-115 in vitro experiments. Cell death was examined by using TUNEL assay on agarose-embedded sections. Then we investigated the effects of TGR5 on PANC-1 and BXPC3 cells via transmission electron microscopy (TEM). Moreover, LC–MS-based metabolomics was performed to explore the potential underlying mechanisms of TGR5 in pancreatic cancer. The correlations between TGR5 and the metabolism-related genes were further analysed by GEPIA 2 database.
Results
We found the proliferation capacities were decreased significantly in PANC-1 and BXPC3 cells after the treatment of SBI-115 for 48 h. The results of TUNEL assay showed that antagonism of TGR5 by SBI-115 had a remarkable effect on inducing cell death. Analysis of TEM demonstrated that SBI-115 treatment could impair the morphology of mitochondria in most PANC-1 and BXPC3 cells. The LC–MS-based analyses revealed that antagonism of TGR5 could alter the metabolic profiles of PANC-1 cells in vitro. Moreover, TGR5 was associated with some metabolism-related genes in pancreatic cancer.
Conclusion
Our data suggests that antagonism of TGR5 may suppress cell proliferation and induce apoptosis in pancreatic cancer cells. TGR5 may affect the metabolism of pancreatic cancer, and TGR5 would be an attractive target for pancreatic cancer treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Bile acids, synthesized from cholesterol in the liver, are metabolized by enzymes produced by intestinal bacteria and are also essential in the maintenance of metabolic homeostasis, insulin sensitivity and innate immunity [1]. Bile acids are highly cytotoxic and regarded as cancer promoters in esophagus, stomach, pancreas, liver and colon. High levels of bile acids in the body predict an increased incidence of tumors originating from these organs [2]. TGR5 is a G protein-coupled bile acid receptor responsive to energy homeostasis, glucose metabolism and bile acid homeostasis [3]. Increasing evidence points to an essential role of TGR5 in responding to bile acid within enterohepatic organs, such as liver, gallbladder and intestine [4]. TGR5 can be activated by bile acids and regulates various signaling pathways such as nuclear factor κB (NF-κB), AKT as well as extracellular signal-regulated kinases (ERK) [5,6,7,8]. Due to the important roles in different signaling pathways, TGR5 becomes a promising target in many diseases. Its agonists may have the potential in the treatment of metabolic and digestive disorders [9, 10]. Moreover, TGR5 is of clinical significance based on the role in carcinogenesis, including gastric cancer, esophageal cancer and hepatocellular carcinoma (HCC) [11,12,13]. Nonetheless, the effect of TGR5 in pancreatic cancer remains controversial. It has been found that TGR5 is expressed at pancreatic acinar cells and acts as an important mediator in bile acid-induced pancreatitis. Previous study also revealed that TGR5 expression was increased in pancreatic cancer tissues compared with the adjacent normal tissues. Meanwhile, TGR5 may be an independent predictor of overall survival in pancreatic cancer patients [14, 15]. Thus, the further deeper research remains of great importance. Therefore, a novel small molecule TGR5 antagonist SBI-115 was utilized to assess the effect of antagonizing TGR5 on pancreatic cancer cells [16].
2 Material and method
2.1 Cell culture
The human pancreatic cancer cell lines PANC-1, ASPC1, BXPC3, CFPAC-1 and MIA PaCa-2 were kindly provided by Department of General Surgery, Zhongshan Hospital, Fudan University and kept as previously reported [17, 18]. All cell lines have been identified and confirmed to be free of mycoplasma contamination before the experiments. Cells were cultured in 5% CO2 and 95% air incubator, with the temperature at 37 ℃.
2.2 Western blot analysis
Total proteins of above cell lines were lysed with RIPA buffer (Biosharp, BL651A) supplemented with 1% PMSF (Beyotime, ST506), and then mixed with loading buffer. After boiling for 10 min at 100 ℃, equal amounts of protein were separated by 10% SDS-PAGE (Beyotime) and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). For blocking the nonspecific binding, the membranes were incubated with 5% skim milk at room temperature for 2 h and then probed with anti-TGR5 antibody (1:1000, Abcam, ab72608) and anti-β-actin (1:2000, Cell Signaling, Rabbit mAb, 4970) overnight at 4℃. After being rinsed with TBS-T (TBS containing 0.05% Tween-20), the PVDF membranes were incubated with HRP-conjugated secondary antibody (1:5000, Abcam, ab6721) for 2 h at room temperature. Afterwards, the proteins were detected by utilizing an enhanced chemiluminescence kit (New Cell & Molecular Biotech Co., Ltd, Suzhou, Jiangsu, China) and β-actin was used as an internal control. To determine the relative quantity of each protein band, ratios of TGR5 proteins/ β-actin were performed using ImageJ software.
2.3 CCK8 assay
In order to find the suitable antagonistic concentrations of each cell line in response to SBI-115, the different final concentrations of SBI-115 (TGR5 antagonist; MCE, HY-111534) were set as 20, 10, 5 and 1 μM by utilizing the CCK8 assay, and an equal amount of dimethyl sulfoxide (DMSO) was added into the control group. The pancreatic cancer cell lines were collected and seeded into 96-well plates with a density of 3 × 104 cells/ml, 100 μl per well for 24 h. Then SBI-115 was added into per well with different final concentrations. Afterwards, the cells were incubated with 10 μl CCK8 solution (Dojindo, Kumamoto, Japan) per well for 2 h at 37℃ at different detection time points. The optical density (OD) was detected with at a wavelength of 450 nm.
2.4 Clonal formation assay
The cells were seeded into the six-well plate at a density of 1000 cells per well in triplicate and followed by incubating in complete culture medium at 37℃ for 15d. The cells were washed with PBS and then fixed with 4% paraformaldehyde for 30 min. Afterwards, 0.1% (w/v) crystal violet was utilized to stain the cells. The number of colonies was counted by using ImageJ software. All assays were performed in triplicate.
2.5 Wound healing assay
The wound healing assay was performed to assess the cell migration capacity. Briefly, the cells were collected and seeded into 6-well plates. When the cells reached 100% confluence, a 200-µl plastic pipette tip was used to make a scratch on the cell monolayer followed by 3 washes in PBS. After removing the cell debris, the serum-free DMEM was added into 6-well plates. The wounds were recorded at 0 h and 48 h. The extent of wound closure was analyzed by using ImageJ software and all assays were performed in triplicate.
2.6 Transwell cell migration and invasion assays
The 12-well transwell chamber (8 µm pore size; Corning, NY, USA) was applied to analyze the migratory and invasive abilities of pancreatic cancer cells. For the transwell cell migration assay, 2 × 105 pancreatic cancer cells in 200 µl serum-free DMEM were directly seeded into the upper chamber, while 800 µl DMEM with 10% FBS was added to the lower chamber. After 24 h incubation, the migrating cells were fixed with 4% paraformaldehyde and stained with crystal violet. For the transwell cell invasion assay, the 12-well transwell chamber was coated with 50 µl of 1:8 mixture of Matrigel (BD Biosciences) and serum-free DMEM for 2 h at 37 ℃. Then the 4 × 105/ml pancreatic cancer cells were seeded into the upper chamber and 800 µl DMEM containing 10% FBS was added to the lower chamber. After 24 h, the cell was fixed and stained followed by imaging at 100 × magnification under an inverted light microscope. The number of cells that migrated and invaded through the chamber filter was counted by the ImageJ software.
2.7 TUNEL assay
To detect the dying cells, the terminal deoxynucleotidyl transferase-mediated uridine 5′-triphosphate-biotin nick-end labeling (TUNEL) assay was performed. Briefly, the cells were collected from the six-well plate and fixed with paraformaldehyde. Afterwards, these cells were gently embedded in the low-melting-point agarose after centrifugation at 1600 rpm for 5 min. Then the agarose-embedded sections were incubated in 0.1% of Triton X at room temperature for 10 min after dehydrating in different gradients of alcohol. Then the TUNEL kit (Sigma, USA) was used for the estimation of apoptosis.
2.8 Transmission electron microscopy
The cells in SBI-115-treated group and DMSO control group were collected after centrifuge. Then TEM fixative was added and let the precipitation re-suspended in the fixative. After a graded ethanol series, the fixed cells were dehydrated and embedded in the pure EMBed 812, and then kept in 37 ℃ overnight. The TEM was performed and took pictures.
2.9 Non-targeted LC–MS metabolic profiling
After treatment with SBI-115 or DMSO (control group) in PANC-1 cells for 48 h, the culture supernatants were collected from SBI-115-treated cells (n = 6) and DMSO-treated cells (n = 6) for LC–MS based non-targeted metabolomics analyses. The detailed protocol was as follows according to Shanghai Lu-Ming Biotech Co., Ltd. 1000 μl supernatants from each sample were added to an 1.5 ml microtube and then dried in a freeze drier. L-2-chlorophenylalanine (0.3 mg/mL) dissolved in methanol as internal standard and the mixture of methanol and water (1/4, vol/vol) were added to each sample. After being extracted by ultrasonic in ice-water and placed at − 20℃ for 2 h, the samples were centrifuged at 13,000 rpm speed for 10 min. The supernatants were collected using crystal syringes and transferred to LC vials for LC–MS analysis. After analyzed by the Nexera UPLC system (Shimadzu Corporation, Japan), the original LC–MS data was processed by software Progenesis QI V2.3 (Nonlinear, Dynamics, Newcastle, UK) for baseline filtering, peak identification, integral, retention time correction, peak alignment, and normalization. The principal component analysis (PCA) and Orthogonal Partial Least-Squares-Discriminant Analysis (OPLS-DA) were utilized to distinguish the metabolites that differed between groups. To prevent overfitting, sevenfold cross-validation and 200 Response Permutation Testing (RPT) were used to evaluate the quality of the model. Variable Importance of Projection (VIP) values obtained from the OPLS-DA model were used to rank the overall contribution of each variable to group discrimination. The fold change (FC), VIP value and P value were used to screen the differential metabolites as follows: VIP ≥ 1 from the cross-validated OPLS-DA models, P value < 0.05 and |log2FC|> 0.25.
2.10 The correlation analyses between TGR5 and the metabolism-related genes by GEPIA 2
To further validate the relationships between TGR5 expression level and the metabolic pathways in pancreatic cancer, the metabolism-related genes were chosen for correlation analyses by GEPIA 2 (http://gepia2.cancer-pku.cn/). GEPIA 2 is an enhanced web server which allows users to analyze gene expression profiles based on tumor and normal samples from the TCGA and the GTEx databases [19]. Gene expression correlation analyses were performed by the Spearman method using the "Correlation" module of GEPIA 2. The “PAAD Tumor”, “PAAD Normal” and “Pancreas” were selected for analysis. The strength of correlation was calculated as (R). TGR5 was represented on the x-axis, and other genes were represented on the y-axis.
2.11 Statistical analysis
Statistical analysis was performed using SPSS 23.0 software (SPSS, Inc., Chicago, IL, USA). The continuous values of experiments were expressed as means ± standard deviation (SD). The differences were analyzed by using Student's t-test when comparing only two groups. The strength of the correlation was determined as follows: very weak (0.00–0.19), weak (0.20–0.39), moderate (0.40–0.59), strong (0.60–0.79), and very strong (0.80–1.0) [20]. P < 0.05.*P < 0.05, **P < 0.01 or ***P < 0.001 were considered statistically significant.
3 Results
3.1 SBI-115 can antagonize the expression of TGR5 at protein level in PANC-1 and BXPC3 cells
Human pancreatic cancer cell lines have different characteristics and remain the biological complexity, such as DNA fingerprinting, functional profiles, et al. [21, 22]. To explore the potential role of TGR5 in pancreatic cancer, its protein level was first examined by western blot in a panel of five human pancreatic cancer (PANC-1, ASPC1, BXPC3, CFPAC-1, MIA PaCa-2). Western blot analyses showed that TGR5 protein levels were upregulated both in PANC-1 and BXPC3 cell lines compared with those in ASPC1, CFPAC-1 and MIA PaCa-2 pancreatic cancer cells (Fig. 1A) (P < 0.01). Based on these results, we performed the follow-up TGR5 antagonism studies with the PANC-1 and BXPC3 cells (TGR5-high) instead of the ASPC1, CFPAC-1 and MIA PaCa-2 cells (TGR5-low). Then the suitable antagonistic concentration values of SBI-115 on PANC-1 and BXPC3 cells were explored by CCK8 assay. After 48 h of SBI-115 treatment, the concentrations resulted in about 50% percent death of PANC-1 and BXPC3 cells were chosen for the subsequent study. The results indicated that SBI-115 exhibited significant inhibitory activities against the PANC-1 and BXPC3 cell lines at 10 μM and 5 μM, respectively (Fig. 1B). In order to validate whether SBI-115 antagonized the expression of TGR5, the western blot assay was applied to detect the protein level of TGR5. We found that TGR5 expressions in PANC-1 and BXPC3 cells were weakened within 48 h after the treatment of SBI-115 at 10 μM and 5 μM, respectively (Fig. 1C) (P < 0.001). These results indicated that SBI-115 can antagonize the expression of TGR5 at protein level in pancreatic cancer cell lines.
SBI-115 can antagonize the expression of TGR5 in pancreatic cancer cells. A The protein levels of TGR5 in five human pancreatic cancer lines (PANC-1, ASPC1, BXPC3, CFPAC-1, MIA PaCa-2); B The effects of different concentrations of SBI-115 on the cell viabilities of PANC-1 and BXPC3 cell lines by using CCK-8; C The result of Western blot showed that TGR5 expressions in SBI-115-treated PANC-1 and BXPC3 groups were weakened compared with that control groups. **P < 0.01, ***P < 0.001
3.2 Pancreatic cell proliferation is inhibited while antagonizing TGR5
The results of cell proliferation by CCK-8 and colony formation showed that the proliferation abilities of SBI-115-treated PANC-1 and BXPC3 cells were decreased compared with the control groups (Fig. 2A, B). However, the results of scratch wound healing assays and transwell cell migration assays indicated that antagonizing TGR5 had no effect on the pancreatic cancer cell migratory capacities (Fig. 2C, D). In addition, the results of transwell cell invasion assays also indicated that antagonizing TGR5 had no effect on the pancreatic cancer cell invasive capacities (Fig. 2E).
Antagonism of TGR5 by SBI-115 suppressed cell proliferation, but had no effects on cell migration and invasion in PANC-1 and BXPC3 cells. A The effects of SBI-115 on cell proliferation by CCK-8 in PANC-1 and BXPC3 cells; B The effects of SBI-115 on cell proliferation by using colony formation assays in PANC-1 and BXPC3 cells; C The effects of SBI-115 on cell migratory abilities by scratch wound healing assays in PANC-1 and BXPC3 cells; D The effects of SBI-115 on cell migratory abilities by transwell assay in PANC-1 and BXPC3 cells; E The effects of SBI-115 on cell invasive capacities by transwell assay in PANC-1 and BXPC3 cells. *P < 0.5, **P < 0.01, ***P < 0.001, ns no significant
3.3 Antagonizing TGR5 induces pancreatic cancer cell apoptosis in vitro
To further explore the oncogenic role of TGR5 in pancreatic cancer, TUNEL assay was used for apoptosis detection.The results indicated that the proportions of TUNEL-positive PANC-1 and BXPC3 cells in SBI-115-treated-groups were higher than those in control groups (Fig. 3).
3.4 The cell morphologies and ultrastructural changes induced by SBI-115 in vitro
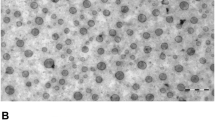
For further research, we treated cells with SBI-115 or DMSO at an initial density of 3 × 104 cells/ml for 48 h in complete culture medium, then the clear visualization of cell morphologies and ultrastructural changes were observed under inverted microscope and transmission electron microscope, respectively. In the control group, human PANC-1 and BXPC3 cells exhibited full body, clear border and close arrangement. Meanwhile, PANC-1 and BXPC3 cells treated with SBI-115 showed unregulated-shaped body and more visible vacuoles (Fig. 4A). Furthermore, the transmission electron microscopy was used to determine the ultrastructural changes in SBI-115-treated cells. It was shown that the mitochondria in SBI-115-treated cells exhibited decreased mitochondrial cristae, swollen-shaped body and ruptured mitochondrial membranes while the mitochondria in DMSO control groups were linear with an integral structure (Fig. 4B).
The cell morphologies and ultrastructural changes induced by SBI-115 in vitro. A The cellular morphologies of PANC-1 and BXPC3 cells were observed by using the light inverted microscope (100 ×); B The ultrastructural changes in PANC-1 and BXPC3 cells were observed by transmission electron microscope (1 μm). The red arrows indicated the mitochondria
3.5 Antagonism of TGR5 alters the metabolic profiles in PANC-1 cells
The LC–MS based non-targeted metabolomics analyses were performed in culture supernatants to compare the metabolic profiling changes between the SBI-115-treated-group and the DMSO-treated group in PANC-1 cells. The PCA and OPLS-DA score plots indicated the great differences between these two groups and the distributions were relative clustered (Fig. 5A, B). Moreover, the values of R2 = 0.994 and Q2 = − 0.036 demonstrated the stable and reliable analytical platform in the OPLS-DA model (Fig. 5C). The S-plot derived from OPLS-DA suggested the accumulated differential metabolites in SBI-115-treated-group and the DMSO-treated group (Fig. 5D). Comparing the metabolites of SBI-115-treated-group versus those of DMSO-treated group, a total of 367 differential metabolites were screened. Among them, 122 differential metabolites were significantly up-regulated while 245 differential metabolites were down-regulated in SBI-115-treated-group when compared with the DMSO-treated group. As shown in volcano map, the blue and red dots represented the significantly down-regulated and up-regulated metabolites in SBI-115-treated-group, respectively (Fig. 6A). Moreover, KEGG pathway enrichment analyses showed the altered metabolites were significantly related to (1) choline metabolism in cancer; (2) tryptophan metabolism; (3) glycerophospholipid metabolism, et al. (Fig. 6B) (Fig. S1A–C). The altered metabolites in above KEGG pathways were represented in Table 1. Fold change (FC) values represented the ratios of average amount of metabolites in SBI-115-treated groups versus DMSO-treated groups.
The LC–MS based non-targeted metabolomics analyses in SBI-115-treated group and DMSO-treated group. A, B The PCA score plot and OPLS-DA score plot showed the differences in metabolic components were significant between the SBI-115-treated group and DMSO-treated group; C The permutation test (200 times) of the OPLS-DA model in the SBI-115-treated group and DMSO-treated group; D The S-plot derived from OPLS-DA demonstrated the accumulated differential metabolites in the SBI-115-treated-group and DMSO-treated group. LC–MS liquid chromatography-mass spectrometry, DMSO dimethyl sulfoxide, PCA principal component analysis, OPLS-DA Orthogonal Partial Least-Squares-Discriminant Analysis
Volcano map of differential metabolites and KEGG pathway enrichment analysis based on the LC–MS metabolomics assay. A The volcano map displayed 122 up-regulated metabolites and 245 down-regulated metabolites in SBI-115-treated-group when compared with the DMSO-treated group. The blue and red dots represented the significantly down-regulated and up-regulated metabolites in SBI-115-treated-group, respectively; B The KEGG pathway enrichment analysis was expressed as a bubble diagram. The size of the each bubble indicated the number of metabolites. The color of bubbles represented different P values. KEGG Kyoto Encyclopedia of Genes and Genomes, LC–MS liquid chromatography–mass spectrometry, DMSO dimethyl sulfoxide
3.6 Relationships between TGR5 and the metabolism-related genes in pancreatic cancer
To further validate the potential role of TGR5 in choline metabolism, tryptophan metabolism and glycerophospholipid metabolism, the correlations between TGR5 and these metabolism-related genes in pancreatic cancer were analyzed. The following genes were identified for subsequent validation: the choline metabolism-related genes, choline transporter-like protein 1 (CTL1), choline transporter 1 (CHT1), choline kinase alpha (CHKA); the tryptophan metabolism-related gene indoleamine 2,3-dioxygenase 1 (IDO1) and the glycerophospholipid metabolism-related gene glycerol-3-phosphate acyltransferase-1 (GPAT1) [23,24,25]. The correlation analyses indicated TGR5 exhibited moderate positive associations with CHT1 and GPAT1 (R = 0.41, P < 0.05; R = 0.41, P < 0.05). Moreover, TGR5 exhibited strong positive associations with CTL1 and IDO1 (R = 0.77, P < 0.05; R = 0.75, P < 0.05) (Fig. 7). These results validated that TGR5 was associated with the choline metabolism, tryptophan metabolism and glycerophospholipid metabolism in pancreatic cancer.
The correlation analyses between TGR5 and the metabolism-related genes in GEPIA 2. TGR5 exhibited moderate positive associations with CHT1 and GPAT1 (R = 0.41, P < 0.05; R = 0.41, P < 0.05), and TGR5 exhibited strong positive associations with CTL1 and IDO1 (R = 0.77, P < 0.05; R = 0.75, P < 0.05). A CTL1; B CTH1; C CHKA; D IDO1; E GPAT1. CTL1 transporter-like protein 1, CTH1 choline transporter 1, CHKA choline kinase alpha, IDO1 indoleamine 2,3-dioxygenase 1, GPAT1 glycerol-3-phosphate acyltransferase-1
4 Discussion
Pancreatic cancer is currently among the most lethal diseases and efficacious therapeutic options are limited [26]. Pancreatic cancer cells adopt metabolic reprogramming to meet the energy demand and result in the carcinogenesis and progression. In addition, metabolic reprogramming is also closely related to chemotherapy, radiotherapy and immunotherapy in pancreatic cancer [27]. Treating cancer by targeting the unique metabolic reprogramming has emerged as a promising therapeutic approach for treatments against pancreatic cancer [28].
Bile acids play an important role in intestinal nutrient absorption, biliary secretion of lipids and toxic metabolites[29]. Moreover, bile acids also participate in regulating energy homeostasis and maintaining metabolic homeostasis as regulatory molecules. Disorders in bile acids synthesis and metabolism may cause metabolic syndrome, inflammation and liver diseases. Bile acids were also identified as a potential carcinogen [2, 30]. Previous studies revealed that bile acids were considered as tumor-promoting factors in the development of human gastrointestinal cancers and elevated levels of bile acids may cause DNA damage [2]. It was also found that bile acids were associated with the risk factors of pancreatic cancer, including alcohol, smoking and a high-fat diet [31, 32]. These risk factors can lead to excess bile acid secretion, which promotes the progression of pancreatitis. Pancreatitis may gradually develop into malignant tumors [33]. Thus, bile acids may be involved in pancreatic cancer carcinogenesis.
TGR5 (also called GPBAR1) is a bile acid-specific cell membrane G protein-coupled receptor linked to cAMP signaling, which participates in bile acids synthesis, energy metabolism and homeostasis. Moreover, previous studies demonstrated that high TGR5 may be a risk factor for cancer, such as cholangiocarcinoma, non-small cell lung cancer (NSCLC). TGR5 overexpression could promote cell proliferation and tumor growth in NSCLC [34,35,36]. It was found that TGR5 was significantly higher in pancreatic cancer tissues than the adjacent normal tissues. High TGR5 suggested poor survival in pancreatic cancer patients [14]. Our data indicated that the proliferation ability of pancreatic cancer cells was decreased significantly when antagonizing TGR5. Meanwhile, antagonism of TGR5 by SBI-115 induced apoptosis in pancreatic cancer cells. However, the significant alterations in the migratory and invasive abilities of SBI-115-treated pancreatic cancer cells were not observed in our study. It was reported that the levels of cyclic adenosine monophosphate (cAMP) and cell proliferation were decreased in cystic cholangiocytes after treatment of SBI-115. On the contrary, TGR5 agonists increased cAMP levels and the ability of cystic cholangiocytes proliferation [16]. Previous study revealed that blocking cAMP-dependent intracellular signalings may prevent the development of pancreatic cancer. It was also found that the development of pancreatic cancer was prevented by cAMP decrease in hamster models [37, 38]. However, few studies have been reported to directly focus on the role of cAMP in migration and invasion of pancreatic cancer cells.
For further study, the cell morphologies and ultrastructural changes induced by SBI-115 in pancreatic cancer cells were further examined. Morphologically, PANC-1 and BXPC3 cells treated with SBI-115 exhibited unregulated-shaped bodies and more visible vacuoles, and the transmission electron microscopy showed the mitochondria in SBI-115-treated cells exhibited decreased mitochondrial cristae, swollen-shaped body and ruptured mitochondrial membranes. Mitochondria are important organelles for cellular physiology and homeostasis in most eukaryotic organisms, and bile acids could directly act on the mitochondria [39]. TGR5 ablation did influence the mitochondrial biogenetic pathway [40]. Our results demonstrated that antagonism of TGR5 would lead to the abnormal structure of mitochondria in pancreatic cancer cells, but further verification was also still needed.
Meanwhile, the LC–MS-based metabolomics analysis revealed that antagonism of TGR5 could alter the metabolic profiles in PANC-1 cells. A total of 122 differential metabolites were significantly up-regulated and 245 differential metabolites were down-regulated in SBI-115-treated-group when compared with the DMSO-treated group. Furthermore, the KEGG enrichment analyses showed the altered metabolites were significantly related to choline metabolism in cancer, tryptophan metabolism, glycerophospholipid metabolism, et al. The correlation analyses between TGR5 and the metabolism-related genes in GEPIA 2 also suggested that TGR5 was significantly correlated with the choline metabolism, tryptophan metabolism and glycerophospholipid metabolism in pancreatic cancer. Previous studies indicated that choline metabolism, tryptophan metabolism and glycerophospholipid metabolism were altered in a wide variety of cancers. Activated choline metabolism is a hallmark of carcinogenesis and tumor progression, leading to elevated levels of phosphocholine and glycerophosphocholine in multiple cancers [41]. Tryptophan metabolism can promote tumor progression by suppressing antitumour immune responses and increasing the malignant properties of cancer cells [24, 42, 43]. They may be promising targets in the treatment of pancreatic cancer [28, 44].
To the end, there are still some shortcomings in our study. First, more animal models are urgently needed to verify the effect of TGR5 in pancreatic cancer. Second, the basic molecular mechanisms underlying TGR5 in pancreatic cancer have been obscure. Third, SBI-115 is a novel TGR5 antagonist and more exploratory studies are still needed by in vivo animal experiments and in vitro cell experiments. In conclusion, our study demonstrates that antagonism of TGR5 suppresses cell proliferation and induces apoptosis in pancreatic cancer cells. TGR5 may be a potential therapeutic target in pancreatic cancer.
Data availability
Derived data supporting the findings of this study are available from the corresponding author on reasonable request.
Abbreviations
- TGR5:
-
Takeda G protein receptor 5
- LC-MS:
-
Liquid chromatography mass spectrometry
- TEM:
-
Transmission electron microscopy
References
Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111–28. https://doi.org/10.1038/nrgastro.2017.119.
Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47–65. https://doi.org/10.1016/j.mrrev.2004.08.001.
Guo C, Chen WD, Wang YD. TGR5, not only a metabolic regulator. Front Physiol. 2016;7:646. https://doi.org/10.3389/fphys.2016.00646.
Reich M, Klindt C, Deutschmann K, Spomer L, Häussinger D, Keitel V. Role of the G protein-coupled bile acid receptor TGR5 in liver damage. Dig Dis. 2017;35:235–40. https://doi.org/10.1159/000450917.
Meng Z, Liu N, Fu X, Wang X, Wang YD, Chen WD, Zhang L, Forman BM, Huang W. Insufficient bile acid signaling impairs liver repair in CYP27(-/-) mice. J Hepatol. 2011;55:885–95. https://doi.org/10.1016/j.jhep.2010.12.037.
Kida T, Tsubosaka Y, Hori M, Ozaki H, Murata T. Bile acid receptor TGR5 agonism induces NO production and reduces monocyte adhesion in vascular endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33:1663–9. https://doi.org/10.1161/ATVBAHA.113.301565.
Masyuk AI, Huang BQ, Radtke BN, Gajdos GB, Splinter PL, Masyuk TV, Gradilone SA, LaRusso NF. Ciliary subcellular localization of TGR5 determines the cholangiocyte functional response to bile acid signaling. Am J Physiol Gastrointest Liver Physiol. 2013;304:G1013–24. https://doi.org/10.1152/ajpgi.00383.2012.
Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–20. https://doi.org/10.1194/jlr.R900007-JLR200.
Kumar DP, Rajagopal S, Mahavadi S, Mirshahi F, Grider JR, Murthy KS, Sanyal AJ. Activation of transmembrane bile acid receptor TGR5 stimulates insulin secretion in pancreatic β cells. Biochem Biophys Res Commun. 2012;427:600–5. https://doi.org/10.1016/j.bbrc.2012.09.104.
Broeders EP, Nascimento EB, Havekes B, Brans B, Roumans KH, Tailleux A, Schaart G, Kouach M, Charton J, Deprez B, Bouvy ND, Mottaghy F, Staels B, van MarkenLichtenbelt WD, Schrauwen P. The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab. 2015;22:418–26. https://doi.org/10.1016/j.cmet.2015.07.002.
Casaburi I, Avena P, Lanzino M, Sisci D, Giordano F, Maris P, Catalano S, Morelli C, Andò S. Chenodeoxycholic acid through a TGR5-dependent CREB signaling activation enhances cyclin D1 expression and promotes human endometrial cancer cell proliferation. Cell Cycle. 2012;11:2699–710. https://doi.org/10.4161/cc.21029.
Carino A, Graziosi L, D’Amore C, Cipriani S, Marchianò S, Marino E, Zampella A, Rende M, Mosci P, Distrutti E, Donini A, Fiorucci S. The bile acid receptor GPBAR1 (TGR5) is expressed in human gastric cancers and promotes epithelial-mesenchymal transition in gastric cancer cell lines. Oncotarget. 2016;7:61021–35. https://doi.org/10.18632/oncotarget.10477.
Nagathihalli NS, Beesetty Y, Lee W, Washington MK, Chen X, Lockhart AC, Merchant NB. Novel mechanistic insights into ectodomain shedding of EGFR Ligands Amphiregulin and TGF-α: impact on gastrointestinal cancers driven by secondary bile acids. Cancer Res. 2014;74:2062–72. https://doi.org/10.1158/0008-5472.CAN-13-2329.
Zhao RY, He SJ, Ma JJ, Hu H, Gong YP, Wang YL, Hu BJ, Xie JZ, Tu WZ, Huang Q, Cheng J. High expression of TGR5 predicts a poor prognosis in patients with pancreatic cancer. Int J Clin Exp Pathol. 2018;11:3567–74.
Muili KA, Jin S, Orabi AI, Eisses JF, Javed TA, Le T, Bottino R, Jayaraman T, Husain SZ. Pancreatic acinar cell nuclear factor κB activation because of bile acid exposure is dependent on calcineurin. J Biol Chem. 2013;288:21065–73. https://doi.org/10.1074/jbc.M113.471425.
Masyuk TV, Masyuk AI, Lorenzo Pisarello M, Howard BN, Huang BQ, Lee PY, Fung X, Sergienko E, Ardecky RJ, Chung TDY, Pinkerton AB, LaRusso NF. TGR5 contributes to hepatic cystogenesis in rodents with polycystic liver diseases through cyclic adenosine monophosphate/Gαs signaling. Hepatology. 2017;66:1197–218. https://doi.org/10.1002/hep.29284.
Yin H, Pu N, Chen Q, Zhang J, Zhao G, Xu X, Wang D, Kuang T, Jin D, Lou W, Wu W. Gut-derived lipopolysaccharide remodels tumoral microenvironment and synergizes with PD-L1 checkpoint blockade via TLR4/MyD88/AKT/NF-κB pathway in pancreatic cancer. Cell Death Dis. 2021;12:1033. https://doi.org/10.1038/s41419-021-04293-4.
Pu N, Gao S, Yin H, Li JA, Wu W, Fang Y, Zhang L, Rong Y, Xu X, Wang D, Kuang T, Jin D, Yu J, Lou W. Cell-intrinsic PD-1 promotes proliferation in pancreatic cancer by targeting CYR61/CTGF via the hippo pathway. Cancer Lett. 2019;460:42–53. https://doi.org/10.1016/j.canlet.2019.06.013.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–60. https://doi.org/10.1093/nar/gkz430.
Liang Y, Lei Y, Liang M, Du M, Liu Z, Li X, Meng X, Zhou B, Gao Y. GBE1 is an independent prognostic marker and associated with CD163+ tumor-associated macrophage infiltration in lung adenocarcinoma. Front Oncol. 2022;11: 781344. https://doi.org/10.3389/fonc.2021.781344.
Satake T, Suetsugu A, Nakamura M, Hasegawa K, Kunisada T, Shimizu M, Saji S, Moriwaki H, Hoffman RM. Differential organ-targeting and cellular characteristics of metastatic human pancreatic cancer cell lines in mouse models. Anticancer Res. 2018;38:1927–35. https://doi.org/10.21873/anticanres.12430.
Chifenti B, Morelli M, Zavaglia M, Coviello DA, Guerneri S, Santucci A, Paffetti A, Masetti M, Locci MT, Bertacca G, Capodanno A, Collecchi P, Campani D, Mosca F, Bevilacqua G, Cavazzana AO. Establishment and characterization of 4 new human pancreatic cancer cell lines: evidences of different tumor phenotypes. Pancreas. 2009;38:184–96. https://doi.org/10.1097/MPA.0b013e31818c746a.
Penet MF, Shah T, Bharti S, Krishnamachary B, Artemov D, Mironchik Y, Wildes F, Maitra A, Bhujwalla ZM. Metabolic imaging of pancreatic ductal adenocarcinoma detects altered choline metabolism. Clin Cancer Res. 2015;21:386–95. https://doi.org/10.1158/1078-0432.CCR-14-0964.
Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov. 2019;18:379–401. https://doi.org/10.1038/s41573-019-0016-5.
Gulvady AA, Murphy EJ, Ciolino HP, Cabrera RM, Jolly CA. Glycerol-3-phosphate acyltransferase-1 gene ablation results in altered thymocyte lipid content and reduces thymic T cell production in mice. Lipids. 2013;48:3–12. https://doi.org/10.1007/s11745-012-3741-7.
Tang R, Zhang Y, Liang C, Xu J, Meng Q, Hua J, Liu J, Zhang B, Yu X, Shi S. The role of m6A-related genes in the prognosis and immune microenvironment of pancreatic adenocarcinoma. PeerJ. 2020;8: e9602. https://doi.org/10.7717/peerj.9602.
Qin C, Yang G, Yang J, Ren B, Wang H, Chen G, Zhao F, You L, Wang W, Zhao Y. Metabolism of pancreatic cancer: paving the way to better anticancer strategies. Mol Cancer. 2020;19:50. https://doi.org/10.1186/s12943-020-01169-7.
Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31:5–19. https://doi.org/10.1016/j.ccell.2016.12.006.
Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191–212. https://doi.org/10.1002/cphy.c120023.
Shen Y, Liu K, Luo X, Guan Q, Cheng L. A simple and reliable bile acid assay in human serum by LC-MS/MS. J Clin Lab Anal. 2022;36: e24279. https://doi.org/10.1002/jcla.24279.
Feng HY, Chen YC. Role of bile acids in carcinogenesis of pancreatic cancer: an old topic with new perspective. World J Gastroenterol. 2016;22:7463–77. https://doi.org/10.3748/wjg.v22.i33.7463.
Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252–61. https://doi.org/10.1053/j.gastro.2013.01.068.
Pinho AV, Chantrill L, Rooman I. Chronic pancreatitis: a path to pancreatic cancer. Cancer Lett. 2014;345:203–9. https://doi.org/10.1016/j.canlet.2013.08.015.
Zhao L, Zhang H, Liu X, Xue S, Chen D, Zou J, Jiang H. TGR5 deficiency activates antitumor immunity in non-small cell lung cancer via restraining M2 macrophage polarization. Acta Pharm Sin B. 2022;12:787–800. https://doi.org/10.1016/j.apsb.2021.07.011.
Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–40. https://doi.org/10.1074/jbc.M209706200.
Li AD, Xie XL, Qi W, Wang WB, Ma JJ, Zhao DQ, Jiang XY, Chen L, Bai Y, Jiang HQ. TGR5 promotes cholangiocarcinoma by interacting with mortalin. Exp Cell Res. 2020;389: 111855. https://doi.org/10.1016/j.yexcr.2020.111855.
Al-Wadei HA, Al-Wadei MH, Schuller HM. Prevention of pancreatic cancer by the beta-blocker propranolol. Anticancer Drugs. 2009;20:477–82. https://doi.org/10.1097/CAD.0b013e32832bd1e3.
Banerjee J, Papu John AM, Al-Wadei MH, Schuller HM. Prevention of pancreatic cancer in a hamster model by cAMP decrease. Oncotarget. 2016;7:44430–41. https://doi.org/10.18632/oncotarget.9790.
Tran QT, Tran VH, Sendler M, Doller J, Wiese M, Bolsmann R, Wilden A, Glaubitz J, Modenbach JM, Thiel FG, de Freitas Chama LL, Weiss FU, Lerch MM, Aghdassi AA. Role of bile acids and bile salts in acute pancreatitis: from the experimental to clinical studies. Pancreas. 2021;50:3–11. https://doi.org/10.1097/MPA.0000000000001706.
Teodoro JS, Machado IF, Castela AC, Amorim JA, Jarak I, Carvalho RA, Palmeira CM, Rolo AP. Chenodeoxycholic acid has non-thermogenic, mitodynamic anti-obesity effects in an in vitro CRISPR/Cas9 model of bile acid receptor TGR5 knockdown. Int J Mol Sci. 2021;22:11738. https://doi.org/10.3390/ijms222111738.
Sonkar K, Ayyappan V, Tressler CM, Adelaja O, Cai R, Cheng M, Glunde K. Focus on the glycerophosphocholine pathway in choline phospholipid metabolism of cancer. NMR Biomed. 2019;32: e4112. https://doi.org/10.1002/nbm.4112.
Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, Jugold M, Guillemin GJ, Miller CL, Lutz C, Radlwimmer B, Lehmann I, von Deimling A, Wick W, Platten M. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. https://doi.org/10.1038/nature10491.
Pilotte L, Larrieu P, Stroobant V, Colau D, Dolusic E, Frédérick R, De Plaen E, Uyttenhove C, Wouters J, Masereel B, Van den Eynde BJ. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2012;109:2497–502. https://doi.org/10.1073/pnas.1113873109.
Dolce V, Cappello AR, Lappano R, Maggiolini M. Glycerophospholipid synthesis as a novel drug target against cancer. Curr Mol Pharmacol. 2011;4:167–75. https://doi.org/10.2174/1874467211104030167.
Acknowledgements
Many thanks to everyone who helped me during the COVID-19.
Funding
This study was supported by grants from the Scientific Research and Development Fund from Zhongshan hospital, Fudan university (2018ZSFZ005).
Author information
Authors and Affiliations
Contributions
Conception and design: YL, GL, XW; administrative support: CW, XW; provision of study materials: SG, GG, CL, YC; collection and assembly of data: YL, JL, ML; data analysis and interpretation: YL, GL, CW, XW; manuscript writing: All authors; All authors reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
12672_2022_504_MOESM1_ESM.docx
Fig. S1. The KEGG pathway maps of choline metabolism, tryptophan metabolism and glycerophospholipid metabolism. The red and blue nodes represented upregulated metabolites and downregulated metabolites in SBI-115-treated-group, respectively. A choline metabolism in cancer; B tryptophan metabolism; C glycerophospholipid metabolism. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lei, Y., Li, G., Li, J. et al. Investigation of the potential role of TGR5 in pancreatic cancer by a comprehensive molecular experiments and the liquid chromatography mass spectrometry (LC–MS) based metabolomics. Discov Onc 13, 46 (2022). https://doi.org/10.1007/s12672-022-00504-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-022-00504-2