Abstract
Genetic variants in CYP19A1, the gene encoding aromatase, have been reported to be associated with circulating estrogen concentrations, a key risk factor for breast cancer. The mechanism underlying this association is still unclear; it has been suggested that some of these variants may alter the expression and/or activity of aromatase. Here we analyzed the expression of intra-tumoral CYP19A1 messenger RNA (mRNA) and the genotypes of rs10046, a well-characterized single nucleotide polymorphism in CYP19A1, in 138 breast cancer patients and 15 breast cancer cell lines. The genotype TT was detected in 36 patients and six cell lines, genotype CT in 55 patients and five cell lines, and genotype CC in 28 patients and four cell lines. We found no evidence for a significant association of CYP19A1 levels with rs10046 genotypes, although expression tended to be higher in tumors and cell lines with the homozygous risk genotype TT. We also found no evidence for a significant association of rs10046 genotypes with breast cancer prognosis. In contrast, high CYP19A1 expression was highly significantly associated with a poor overall, disease-free, and metastasis-free survival in estrogen receptor-positive but not negative breast cancer patients. Moreover, CYP19A1 mRNA was significantly elevated in postmenopausal patients and in patients older than 50 years, and a trend towards a positive correlation with ER status and ESR1 mRNA expression was observed. These findings highlight the key role of aromatase in estrogen receptor-positive breast cancer biology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
CYP19A1, a member of the cytochrome P450 gene superfamily, encodes the enzyme aromatase, which catalyzes the last, rate-limiting step in estrogen biosynthesis. By sequential hydroxylation, removal of the axial C-19 methyl group, and aromatization of the A-ring, aromatase converts the androgens testosterone and androstenedione to estradiol and estrone, respectively [1]. Lifetime exposure to elevated concentrations of circulating estrogen is an established risk factor for breast cancer, as are aspects of physiology and lifestyle associated with these elevated levels, such as early menarche, late menopause, nulliparity, high BMI, oral contraceptives, and hormone replacement therapy [2,3,4,5]. Estrogen biosynthesis takes place primarily in the ovaries of premenopausal women but switches to extra-gonadal sites such as adipose tissue, skin, and muscle after menopause, concomitant with the sites of highest aromatase expression [1]. Aromatase is frequently overexpressed by breast tumor and adjacent stroma cells [6, 7].
Aromatase inhibitors such as anastrozole, letrozole, and exemestane are highly effective endocrine therapies of postmenopausal, estrogen receptor (ER)-positive breast cancer, prolonging the disease-free survival (DFS) and overall survival (OS) [8,9,10]. Conversely, high expression of aromatase has been associated with a shortened survival, albeit inconsistently [11,12,13,14,15,16,17,18]. Unfortunately, only few of these studies have specifically analyzed ER-positive patients [16, 19].
Genetic variants of CYP19A1 have been associated with elevated circulating estrogen levels in pre- and postmenopausal women and also in men [3, 20,21,22,23]. Per risk allele, the estradiol/testosterone ratio was found increased by about 10%, which explains ~ 1.1–1.6% of its variance in postmenopausal women [3, 23]. By comparison, the BMI accounts for ~ 16% of the variance of circulating estrogen levels, and the estimated overall heritability is about 40% [23, 24]. Since elevated levels of circulating estrogen increase the risk of breast cancer and other hormone-dependent cancers, it seems plausible that CYP19A1 variants associated with elevated estrogen levels should also be associated with an increased risk of these cancers. However, whereas such an association has been well-established for endometrial cancer [23], most CYP19A1 candidate SNP studies did not find an association with breast cancer risk [25,26,27,28,29,30,31]. This may be due to differences in effect size: the odds ratio associated with a doubling of circulating estradiol levels has been estimated to be 2.06 (95% CI, 1.47–2.89) for endometrial cancer, but only 1.29 (95% CI, 1.15–1.44) for breast cancer [2, 32].
The potential association of CYP19A1 variants with breast cancer prognosis has also been investigated in detail. The CC genotype of rs10046, one of the most extensively studied CYP19A1 candidate SNPs, was found associated with a longer disease-free survival of premenopausal, but not postmenopausal breast cancer patients [33]. Likewise, decreased OS and DFS were found for the minor allele of rs700519, which is in linkage disequilibrium with rs10046, plus five other CYP19A1 SNPs [34]. Another CYP19A1 haplotype was associated with a poor OS, DFS, and metastasis-free survival (MFS) in premenopausal, ER-positive breast cancer patients [35]. In contrast, two other large studies found no evidence for an association of rs10046 and OS, DFS, and MFS in unselected breast cancer patients [36, 37].
The aim of our study was to analyze the association of CYP19A1 messenger RNA (mRNA) expression and the common SNP rs10046 with prognosis specifically in ER-positive vs. ER-negative breast cancer patients. Moreover, we wanted to investigate the association of CYP19A1 expression with widely used clinical and histopathological characteristics of breast cancer, in particular the ER status and the age at onset. Finally, by determining the association of rs10046 genotypes with CYP19A1 expression, we addressed a proposed mechanism by which genetic variants in CYP19A1 might affect circulating estrogen levels and (breast) cancer risk.
Patients and Methods
Study Population
This study was approved and is annually reviewed by the Institutional Review Board (IRB, “Ethikkommission”) of the Medical University of Vienna, Austria (MUV; protocol 141/2002). Written informed consent was obtained from patients recruited after the onset of the study. For those patients who had undergone surgery before the onset of the study, a waiver of specific informed consent was approved by the IRB. One hundred and thirty-eight consecutive breast cancer patients treated between 1991 and 1994 at the Department of Obstetrics and Gynecology, MUV, were enrolled in this study. Detailed follow-up records and fresh-frozen tumor tissue were available for each patient. Only women of Western European descent from the same geographical area were included. Clinical and histopathological characteristics of the study population are shown in Supplementary Table 1. Molecular subtypes were defined based on IHC analyses as follows: Triple negative, ER−, PR−, and HER2−; HER2-enriched, ER−, PR−, and HER2+; Luminal A, ER+ and/or PR+, HER2−; and Luminal B, ER+ and/or PR+, HER2+. None of the patients received any neoadjuvant therapy, or any treatment with aromatase inhibitors, or any treatment prior to tumor tissue isolation. Thiry-five patients received adjuvant chemotherapy, 39 received tamoxifen anti-hormonal therapy, 37 received both, and 21 received no systemic therapy. For six patients, the records are incomplete. Of the 72 patients with chemotherapy, the schemes were CMF (n = 36), modified CMF (n = 9), or other (n = 27). Sixty-one patients were ER positive. Forty-seven of those were treated with tamoxifen, and 14 were not. Of the latter, four received chemotherapy, and ten no systemic therapy.
DNA Isolation and Genotyping
Genomic DNA from 122 patients has been extracted previously from fresh-frozen primary tumor samples with the High Pure PCR Template Preparation Kit (Roche, Vienna, Austria) as described [38]. Genotyping of SNP rs10046 (CYP19 E10 c.+19C>T) was performed by TaqMan PCR with Genotyping Master mix and allele-specific, fluorescently labeled probes on a 7500 fast instrument following the manufacturer’s instructions (Applied Biosystems, Brunn/Gebirge, Austria; Assay-ID # C___8234731_30). Twenty nanograms of genomic DNA was used per reaction in a reaction volume of 10 μl.
qRT-PCR Analysis
Isolation of total RNA from 111 fresh-frozen tumor samples with TRIreagent (Sigma), quality control with the Bioanalyser 2100 (Agilent), and Reverse Transcription with the High-Capacity complementary (cDNA) Archive Kit (Applied Biosystems, Brunn/Gebirge, Austria) has been described previously [38, 39]. For 17 patients, CYP19A1 mRNA levels were also determined in one lymph node metastasis each in addition to the primary tumor. In each of our qRT-PCR runs, which were conducted in 96-well plates, two to four negative controls (2.5 μl ddH2O instead of cDNA) were included and run in parallel. No signal was obtained in any of these reactions. As a positive control, duplicate samples of serial dilutions of a cDNA standard (cultured normal breast epithelial cells; HMECs) were included in each run. Each sample was analyzed in duplicate by quantitative reverse transcription PCR (qRT-PCR) on an Applied Biosystems 7500 fast real-time PCR instrument, using the following gene-specific primers and fluorescent probes obtained from Applied Biosystems: CYP19A1 (aromatase), hs_00903413; ESR1 (estrogen receptor 1; ERα), hs_00174860; CDH1 (E-cadherin), hs_00170423; and ACTB1 (β-actin; control), hs_99999903. The mRNA levels of CYP19A1, ESR1, and CDH1 were normalized to those of ACTB1 in each sample and were further normalized to controls by setting the mean level of four samples of normal breast tissue to unity (1) and expressing the levels of all other samples relative to those. For expression analyses in breast cancer cell lines, four non-cancer cell lines were used as normalization controls (HMEC, Hs578Bst, MCF10A, and MCF10F). Thus determined CDH1 mRNA levels in breast cancer cell lines were assigned to a positive or negative CDH1 status by setting the mean level of the non-cancer cell lines HMEC, MCF10A, and MCF10F to unity and defining cell lines with at least one tenth that level as positive and all others as negative. Of note, we found a large difference of at least 75-fold between CDH1-positive and CDH1-negative cell lines (data not shown). All relative mRNA expression levels are presented as − ΔΔCt values (i.e., as log(2) values) as described [38].
Cell Lines
Finite lifespan untransformed human mammary epithelial cells (HMEC) were kindly provided by M. R. Stampfer and grown in MEGM medium [40]. All other cell lines were purchased from DSMZ (“Deutsche Sammlung von Mikro-Organismen und Zellkulturen”) or ATCC (American Type Culture Collection) and were grown at 37 °C, 5% CO2, and 100% humidity as described [41]. DSMZ and ATCC authenticate all cell lines by STR profiling and other methods before distribution. Total RNA and genomic DNA were isolated from all cell lines within three to eight passages after receipt as described previously [41, 42].
Statistical Analyses
Statistical analyses were performed with R version 3.2.3 (“Wooden Christmas-Tree”), an open-source language and environment for statistical computing [43]. Differences between subgroups of patients or cell lines with respect to relative CYP19A1 mRNA levels were analyzed by unpaired, two-sided t tests unless indicated otherwise. All P values shown are two-sided. P < 0.05 was considered statistically significant.
Survival Analyses
Kaplan-Meier plots were computed with the R survival package [43]. P values to Kaplan-Meier curves were calculated by log-rank tests as described [44]. “Events” were defined as breast-cancer related death in the overall survival, and as affirmation of a distant metastasis, a second primary breast tumor, or a recurrent primary tumor in the disease-free survival. In the analyses of metastasis-free survival, the occurrence of a distant metastasis was considered as an event, and in the analyses of lung-, liver-, and bone-metastasis-free survival, only metastases to the respective target organ were considered. Among the 100 patients for whom expression of CYP19A1 could be determined, 57 events in the overall survival, 62 in the disease-free survival, and 52 in the metastasis-free survival were observed. The mean follow-up times were 7.6 ± 5.1 years for the overall survival (median, 6.7 years; range, 0–14.7 years), 5.5 ± 5.3 years for the disease-free survival (median, 3.4 years; range, 0–14.7 years), and 6.3 ± 5.4 years for the metastasis-free survival (median, 4.4 years; range, 0–14.7 years). For those patients who were still event-free at the end of follow-up, the mean follow-up times were 11.0 ± 4.8 years (median, 13.1 years; range, 0–14.7 years) for the overall survival, 10.1 ± 5.2 years (median, 12.8 years; range, 0–14.7 years) for the disease-free survival, and 9.9 ± 5.2 years for the metastasis-free survival (median, 12.8 years; range, 0–14.7 years). Details for all subgroups (ER positive and negative; CYP19A1 high and low; and rs10046 genotypes) and for all survival analyses are provided in Supplementary Tables 2 and 3.
Results
Association of CYP19A1 mRNA Expression with rs10046 SNP Genotype and ER Status
Genotypes of SNP rs10046 (CYP19 E10 c.+19C>T), located in the 3′UTR in exon 10 of the CYP19A1 gene, were successfully determined in 119 breast cancer patients. The study population exhibited a frequency of 53.4% for the T-allele and was in Hardy-Weinberg equilibrium with respect to rs10046 genotypes (P = 0.52). Clinical characteristics and genotypes of these patients are shown in Supplementary Table 1. rs10046 was also genotyped in 15 human breast cancer cell lines, and genotype TT was detected in six (Cama1, Hcc1937, Hs578T, MDA-MB-231, MDA-MB-468, and ZR75-1), genotype CT in five (AU565, Hcc1143, MDA-MB-435, MDA-MB-453, and SKBR3), and genotype CC in four cell lines (BT474, Kpl1, MCF7, and T47D). CYP19A1 mRNA expression levels were also determined for these 15 cell lines and 100 tumor samples. Overall, CYP19A1 levels were not significantly associated with rs10046 genotypes in breast tumors or breast cancer cell lines (P = 0.256 and P = 0.061, respectively; ANOVA; Fig. 1a, b). However, mean CYP19A1 mRNA levels in samples with the TT genotype were considerably higher than those in CC samples both in tumors (1.9-fold; P = 0.11, t test; Fig. 1a) and in cell lines (9.2-fold; P = 0.01; Fig. 1b).
Association of CYP19A1 mRNA expression with a, b rs10046 SNP genotype, c, d ER (estrogen receptor) status, and e, f ESR1 (estrogen receptor 1) mRNA expression. Analyses are in human primary breast tumors (a, c, e) and in human breast cancer cell lines (b, d, f). CC, CT, and TT are the genotypes of SNP rs10046. The numbers of patients and cell lines in each group (n) are shown in parentheses. The y-axes in a–f show normalized relative CYP19A1 mRNA levels (log(2) values). P values (P, in parentheses above each panel) were determined by a, b ANOVA, c, d unpaired, two-sided t tests, and e, f Spearman’s rank correlation. r, Spearman’s rho (rank correlation coefficient)
Next, potential associations of CYP19A1 expression with estrogen receptor (ER) status were investigated (Fig. 1c, d). Whereas no significant association was found in breast tumor samples (P = 0.337, t test; Fig. 1c), ER-positive breast cancer cell lines exhibited 5.1-fold lower mean CYP19A1 mRNA levels than ER-negative ones (P = 0.050; Fig. 1d). Correlations of CYP19A1 with ESR1 (estrogen receptor 1) mRNA levels were assessed using Spearman’s rank correlation coefficient and visualized by scatterplots (Fig. 1e, f). No such correlation was observed in tumors (r = 0.157, P = 0.121; Fig. 1e). In contrast, a significant negative correlation of CYP19A1 and ESR1 levels was found in tumor cell lines (r = −0.639, P = 0.012; Fig. 1f).
Analysis of CYP19A1 Expression in Human Breast Cancer and Non-cancer Cell Lines
We quantified CYP19A1 mRNA expression levels in 15 human breast cancer cell lines and in four untransformed breast epithelial cell lines (Fig. 2). Breast cancer cell lines tended to have 4.5-fold lower mean CYP19A1 expression levels compared to normal, untransformed breast epithelial cell lines (P = 0.06, t test). In an exploratory analysis, we found that this difference resulted mostly from cell lines with an epithelial morphology and a positive E-cadherin status (Fig. 2). Conversely, E-cadherin-negative tumor cell lines with a fibroblastoid morphology expressed CYP19A1 at roughly the same level as untransformed cell lines. Accordingly, fibroblastoid breast cancer cell lines exhibited a 5.9-fold higher mean CYP19A1 expression compared to cell lines with an epithelial phenotype (P = 0.006; Fig. 2). The ER-positive cell lines within this panel (BT474, Cama1, Kpl1, MCF7, T47D, and ZR75-1) all exhibited an epithelial morphology except Cama1. The five ER-positive epithelial cell lines were among those with the lowest CYP19A1 expression (Figs. 1d and 2).
CYP19A1 mRNA expression in human untransformed breast epithelial and breast cancer cell lines. The y-axis shows mean relative CYP19A1 mRNA levels (log(2) values) of duplicate measurements. Expression levels of the indicated individual untransformed cells (Normal, black bars), breast cancer cell lines with an epithelial morphology (Epithelial, white bars), and breast cancer cell lines with a fibroblastoid morphology in culture (Fib, hatched bars) are shown. For normalization, the mean expression level of the four untransformed cell lines was set to unity (i.e., 20 in graph), and the levels of individual cell lines are presented relative to those. Boxes underneath the bar graph indicate a positive (black) or negative (white) status of ER, PR, and CDH1 (E-cadherin). ER and PR status are based on [45, 46], and CDH1 status is based on determination of mRNA levels by qRT-PCR. Gray, status not available
Association of CYP19A1 Expression with Clinical and Histopathological Characteristics in Primary Human Breast Tumor Tissue
Relative CYP19A1 mRNA expression levels were quantified in primary tumor tissue samples of 100 breast cancer patients. Associations of CYP19A1 expression with well-established clinical and histopathological characteristics of breast cancer were visualized with boxplots (Fig. 3). For 17 of these patients, one lymph node metastasis each was also analyzed, which exhibited 1.5-fold lower mean CYP19A1 levels than their matched primary tumors; however, this difference was not significant (P = 0.113; Fig. 3h). Mean CYP19A1 expression levels were found to be significantly elevated in breast tumor tissue samples of patients with an age at breast cancer onset ≥ 50 years (1.9-fold; P = 0.007; Fig. 3a) and in postmenopausal patients (1.7-fold; P = 0.047; Fig. 3b). In contrast, no significant association with CYP19A1 expression was found in analyses of the tumor type, size, stage, and grade, as well as for lymph node status, p53-, PR-, HER2-status, and the molecular subtype (Fig. 3c–g, i–l), consistent with previous reports [13, 15, 19, 47,48,49,50]. Interestingly, a positive p53 status tended to be associated with both a lower expression of CYP19A1 (Fig. 3i) and a higher frequency of the rs10046 CC genotype (Supplementary Table 1).
Association of CYP19A1 mRNA expression with established clinical and histopathological parameters. Boxplots according to a age at breast cancer onset in years, b menopausal status, c tumor type, d tumor size according to TNM classification, e tumor stage, f tumor grade, g lymph node status, h in paired primary tumors and lymph node metastases (LN) of 17 patients, i p53 status, j progesterone receptor (PR) status, k HER2-status, and l molecular subtype, i.e., HER2-type (HER), Luminal A (Lu.A), Luminal B (Lu.B), and triple negative (TN). neg, negative; pos, positive. The numbers of patients in each group (n) are shown in parentheses. The y-axes show normalized relative CYP19A1 mRNA levels (log(2) values). P values (P, in parentheses above each panel) were determined by e, l ANOVA; h paired, two-sided t tests; and unpaired, two-sided t tests in all other panels
Association of CYP19A1 Expression with Breast Cancer Prognosis
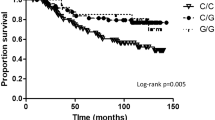
Patients were divided into two subgroups based on CYP19A1 expression by applying maximally selected log rank statistics to define a cutoff in unselected patients as described [44]. Subsequently, Kaplan-Meier analyses of the overall survival (OS), disease-free survival (DFS), and metastasis-free survival (MFS) were performed in unselected patients (Fig. 4a–c), in ER-positive patients (Fig. 4d–f), and in ER-negative patients (Fig. 4g–i). Unselected patients with a high CYP19A1 expression exhibited a significantly decreased MFS (P = 0.026; Fig. 4c), as well as a trend towards a decreased OS (P = 0.115; Fig. 4a) and DFS (P = 0.082; Fig. 4b) compared to patients with a low CYP19A1 expression.
Association of CYP19A1 mRNA expression with the survival of human breast cancer patients. Kaplan-Meier analyses of the overall survival (a, d, g), disease-free survival (b, e, h) and metastasis-free survival (c, f, i) in unselected patients (a–c; n = 100), ER-positive patients (d–f; n = 40), and ER-negative patients (g–i; n = 56) are shown. Patient subgroups with high and low CYP19A1 expression and their numbers (n) are indicated in each panel. ER, estrogen receptor
In ER-positive patients, highly significant differences between patients with high vs. low CYP19A1 expression were observed with respect to OS (P = 3.1 × 10−5; Fig. 4d), DFS (P = 2.1 × 10−4; Fig. 4e), and MFS (P = 1.5 × 10−6; Fig. 4f). In each of these analyses, patients with a high CYP19A1 expression exhibited a very poor survival. All ER-positive patients with a high CYP19A1 expression (n = 11) progressed to metastasis and/or recurrent disease within less than 8 years after diagnosis (Fig. 4e, f). In contrast, ER-negative patients exhibited no difference in OS, DFS, or MFS between high and low CYP19A1 expressers (Fig. 4g–i), indicating that the shorter OS, DFS, and MFS of unselected patients with high CYP19A1 expression results exclusively from the ER-positive subgroup. Adjuvant tamoxifen therapy affected the association of CYP19A1 expression with the survival of these patients more modestly than ER status (Supplementary Fig. 6).
In a hypothesis-generating analysis, metastases to the major target organs lung, liver, and bone were further examined (Supplementary Fig. 1; Supplementary Table 3). Unselected patients with high CYP19A1 expression showed a trend towards decreased lung-MFS (P = 0.068) and bone-MFS (P = 0.072) compared to patients with low CYP19A1 expression, whereas no association was found in the analysis of liver-MFS (P = 0.605; Supplementary Fig. 1). ER-positive patients with high CYP19A1 expression exhibited a very poor survival free of lung metastases (P = 6.3 × 10−7) and bone metastases (P = 1.7 × 10−8) and a significant difference in liver-MFS (P = 0.003; Supplementary Fig. 1). Specifically, all uncensored ER-positive patients with a high CYP19A1 expression (n = 11) had developed both lung and bone metastases within 10 years after diagnosis (Supplementary Fig. 1). CYP19A1 expression was not associated with lung- and bone-MFS in ER-negative patients. In contrast to all other results of our survival analyses, high CYP19A1 expression was associated with a good prognosis with respect to liver-MFS; no liver metastases were observed in this subgroup of 14 patients up to 14 years postdiagnosis (P = 0.043; Supplementary Fig. 1). However, it should be noted that these analyses are limited by the smaller number of events compared to the analyses of OS, DFS, and MFS (Supplementary Tables 2 and 3).
Association of rs10046 SNP Genotypes with Breast Cancer Prognosis
The overall survival (OS), disease-free survival (DFS), and metastasis-free survival (MFS) associated with the three rs10046 genotypes CC, CT, and TT were compared in Kaplan-Meier analyses of unselected patients, ER-positive patients, ER-negative patients, premenopausal patients, and postmenopausal patients (Supplementary Figs. 2 and 4; Supplementary Table 2). No significant differences in any of these analyses were observed. Metastasis to the major target organs lung, liver, and bone was further examined (Supplementary Fig. 3; Supplementary Table 3). The survival analyses in Supplementary Figs. 2 and 3 revealed that patients with the TT genotype tended to have a better prognosis than CC patients if they were ER positive, but a poorer prognosis if they were ER negative. This was most pronounced in the metastases-free survival of ER-negative patients, where the TT genotype was associated with a considerably higher frequency of events than the CC genotype, and the CT genotype was intermediate (Supplementary Figs. 2 and 3; Supplementary Table 2). However, none of these trends was significant. Likewise, patients with the TT genotype tended to have a better prognosis than CC patients if they had received adjuvant tamoxifen therapy, but a poorer prognosis if they were not treated with tamoxifen (Supplementary Fig. 5).
Discussion
Breast tumor cells and/or their surrounding stroma frequently express elevated levels of aromatase, producing sufficient local estrogen concentrations to sustain tumor cell proliferation and progression [6, 7]. Moreover, aromatase inhibitors are a current first line endocrine therapy for postmenopausal, hormone receptor-positive breast cancer [9, 10]. Accordingly, the potential impact of aromatase expression and genetic variants in CYP19A1 on levels of circulating estrogen, breast cancer risk and prognosis, and response to endocrine therapy has been extensively studied [7, 30, 51, 52]. Among other CYP19A1 variants, the rs10046 SNP has been associated with increased levels of circulating estrogen [3, 20,21,22,23] and an increased breast cancer risk in some studies [25, 28]. However, most studies, including the larger ones and meta-analyses, did not find an association of rs10046 with breast cancer risk [26, 27, 29,30,31].
We observed a very poor overall, disease-free, and metastasis-free survival of ER-positive breast cancer patients with a high CYP19A1 expression. Whereas high levels of significance were obtained in ER-positive patients, aromatase expression was not significantly associated with survival in ER-negative patients. Since aromatase functions in the biosynthesis of estrogen, which exerts its breast cancer-promoting effects primarily via the estrogen receptor, these findings appear biologically plausible [5, 52, 53]. Moreover, since aromatase inhibitors prolong the disease-free and overall survival of ER-positive breast cancer, it seems reasonable that high aromatase expression has the opposite effect [8, 9]. A shortened disease-free survival of unselected breast cancer patients with a high CYP19A1 mRNA expression has been reported previously [15]. Moreover, higher CYP19A1 mRNA levels were significantly associated with the incidence of metastasis and local recurrence as well as breast cancer-related death [15]. Consistent with our results, aromatase IHC positivity was associated with a shortened overall survival in ER-positive but not ER-negative breast cancer [16].
However, other studies found no evidence for a significant association of aromatase mRNA, protein, or activity with prognosis in unselected [11,12,13,14, 17] or in ER-positive patients [19]. Intriguingly, low intra-tumoral CYP19A1 mRNA expression was significantly associated with an increased locoregional recurrence rate in premenopausal patients younger than 40 years [18]. These authors suggested that, since estrogen represses the CYP19A1 promoter, CYP19A1 mRNA levels might be inversely correlated with the high circulating estrogen levels in premenopausal patients, consistent with the findings by us and others that CYP19A1 levels are lower in premenopausal patients and in patients younger than 50 years [14, 54]. Accordingly, high estrogen levels might be causal to both low CYP19A1 expression and a high recurrence rate in these premenopausal patients [18]. Three studies have shown that in contrast to low expression, the complete absence of CYP19A1 protein or mRNA expression was associated with a poor prognosis [17, 50, 54]. These seemingly paradoxical results have been suggested to possibly be due to prior adjuvant or neoadjuvant treatment with aromatase inhibitors and/or tamoxifen or to reflect ongoing progression of the disease to a hormone-independent state [17, 50, 54]. We found that a low CYP19A1 expression as well as the TT genotype of rs10046 tended to be associated with a favorable prognosis in patients with, but not those without adjuvant tamoxifen therapy (Supplementary Figs. 5 and 6). However, the number of ER-positive, untreated patients was too small to discern the potential effects of ER status and tamoxifen therapy.
In contrast to CYP19A1 expression, we found no significant association of rs10046 genotypes with breast cancer prognosis in our analyses of the overall survival, disease-free survival, and metastasis-free survival in unselected, ER-positive, and ER-negative patients (Supplementary Figs. 2 and 3). Previous studies have also found either no association or an association in specific subgroups, specifically of the DFS in premenopausal patients [33,34,35,36,37]. In contrast, we did not find a significant association in premenopausal patients. This may in part be due to small numbers, since there were only 33 premenopausal patients with confirmed rs10046 genotypes in our study population (Supplementary Fig. 4).
Like other CYP19A1 variants, the rs10046 SNP has been associated with circulating estrogen levels and the estradiol/testosterone ratio [3, 20,21,22,23]. Per T-allele of rs10046, the estradiol/testosterone ratio was found increased by ~ 10% [3, 23]. rs10046 is located in the 3′ UTR of CYP19A1 and might regulate mRNA stability by miRNA binding and/or other mechanisms. Alternatively, rs10046 could be in strong linkage disequilibrium with one or more causal promoter SNPs rather than having a direct causal effect itself. In a recent systematic analysis, one of several CYP19A1 SNPs other than rs10046, but in linkage disequilibrium with it, was demonstrated to indeed be causal to the elevated estrogen levels [23]. One study has reported that the T-allele of rs10046 is associated with elevated levels of CYP19A1 mRNA in breast tumors [25]. However, that study used a now out-dated method of RNA quantification and did not report actual mRNA levels. Instead, the samples were grouped according to CYP19A1 levels above and below median and subjected to chi-square tests [25]. We are not aware of any additional, more recent analyses of the association of rs10046 genotype with aromatase expression in breast cancer. Here, we found no evidence for a significant association of CYP19A1 levels with rs10046 genotypes in breast tumors or breast cancer cell lines (Fig. 1). However, CYP19A1 levels were clearly elevated in tumors and cell lines with the TT genotype compared to CC samples, in line with the increased circulating estrogen levels associated with the T-allele.
A trend towards a positive correlation between aromatase and ER expression has been reported [14, 15, 49, 50, 55, 56], whereas others found no evidence for a correlation [18, 54, 57,58,59] or even a non-significant inverse correlation [60]. We also found non-significant positive correlations of CYP19A1 mRNA expression with ER status and with ESR1 mRNA expression in breast tumors. In contrast, we found significant inverse correlations in breast cancer cell lines (Fig. 1). However, this in vitro result may be confounded by the fact that five out of six ER-positive cell lines were of epithelial morphology, which was associated with a much lower CYP19A1 expression than the fibroblastoid morphology (Fig. 2). Moreover, it may be explained by culture conditions and by the absence of stromal cells from in vitro culture, which are engaged in extensive gene-regulatory signaling crosstalk with tumor cells in situ and are also a substantial source of intra-tumoral aromatase expression [1, 7].
A possible limitation of the present study relates to the size of our study population. In particular, the number of patients in which both CYP19A1 expression and rs10046 genotype were successfully determined was only 81, which may explain why we were unable to replicate the significantly elevated CYP19A1 mRNA levels in TT patients in a previous study of 99 patients [25]. On the other hand, survival analyses represent the main part of our study, in which the statistical power is determined by the number of events, ameliorating the limitations of our study population. Since other CYP19A1 polymorphisms which are even located within promoter elements were found either not significantly associated [61], or only nominally associated with aromatase expression in non-malignant tissue [23], we believe that the next urgent issue is to convincingly demonstrate the mechanism by which CYP19A1 variants cause the elevated levels of circulating estrogen, be it by altering the expression or the activity of aromatase.
In summary, we report that CYP19A1 mRNA expression is a powerful prognostic marker in ER-positive but not ER-negative breast cancer, is significantly elevated in postmenopausal patients and in patients older than 50 years, and tends to positively correlate with rs10046 risk allele, ER status, and ESR1 mRNA expression.
References
Subramanian A, Salhab M, Mokbel K (2008) Oestrogen producing enzymes and mammary carcinogenesis: a review. Breast Cancer Res Treat 111:191–202. https://doi.org/10.1007/s10549-007-9788-0
Key T, Appleby P, Barnes I, Reeves G (2002) Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J Natl Cancer Inst 94:606–616
Dunning AM, Dowsett M, Healey CS, Tee L, Luben RN, Folkerd E, Novik KL, Kelemen L, Ogata S, Pharoah PD, Easton DF, Day NE, Ponder BA (2004) Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst 96:936–945
Kaaks R, Rinaldi S, Key TJ, Berrino F, Peeters PH, Biessy C, Dossus L, Lukanova A, Bingham S, Khaw KT, Allen NE, Bueno-de-Mesquita HB, van Gils CH, Grobbee D, Boeing H, Lahmann PH, Nagel G, Chang-Claude J, Clavel-Chapelon F, Fournier A, Thiebaut A, Gonzalez CA, Quiros JR, Tormo MJ, Ardanaz E, Amiano P, Krogh V, Palli D, Panico S, Tumino R, Vineis P, Trichopoulou A, Kalapothaki V, Trichopoulos D, Ferrari P, Norat T, Saracci R, Riboli E (2005) Postmenopausal serum androgens, oestrogens and breast cancer risk: the European prospective investigation into cancer and nutrition. Endocr Relat Cancer 12:1071–1082. https://doi.org/10.1677/erc.1.01038
Patel S (2017) Disruption of aromatase homeostasis as the cause of a multiplicity of ailments: a comprehensive review. J Steroid Biochem Mol Biol 168:19–25. https://doi.org/10.1016/j.jsbmb.2017.01.009
Bulun SE, Lin Z, Imir G, Amin S, Demura M, Yilmaz B, Martin R, Utsunomiya H, Thung S, Gurates B, Tamura M, Langoi D, Deb S (2005) Regulation of aromatase expression in estrogen-responsive breast and uterine disease: from bench to treatment. Pharmacol Rev 57:359–383. https://doi.org/10.1124/pr.57.3.6
Wang X, Simpson ER, Brown KA (2015) Aromatase overexpression in dysfunctional adipose tissue links obesity to postmenopausal breast cancer. J Steroid Biochem Mol Biol 153:35–44. https://doi.org/10.1016/j.jsbmb.2015.07.008
Smith IE, Dowsett M (2003) Aromatase inhibitors in breast cancer. N Engl J Med 348:2431–2442. https://doi.org/10.1056/NEJMra023246
Lonning PE (2011) The potency and clinical efficacy of aromatase inhibitors across the breast cancer continuum. Ann Oncol 22:503–514. https://doi.org/10.1093/annonc/mdq337
Harbeck N, Gnant M (2017) Breast cancer. Lancet 389:1134–1150. https://doi.org/10.1016/S0140-6736(16)31891-31898
Lipton A, Santen RJ, Santner SJ, Harvey HA, Sanders SI, Matthews YL (1992) Prognostic value of breast cancer aromatase. Cancer 70:1951–1955
Evans TR, Rowlands MG, Silva MC, Law M, Coombes RC (1993) Prognostic significance of aromatase and estrone sulfatase enzymes in human breast cancer. J Steroid Biochem Mol Biol 44:583–587
Miyoshi Y, Ando A, Hasegawa S, Ishitobi M, Taguchi T, Tamaki Y, Noguchi S (2003) High expression of steroid sulfatase mRNA predicts poor prognosis in patients with estrogen receptor-positive breast cancer. Clin Cancer Res 9:2288–2293
Zhang Z, Yamashita H, Toyama T, Omoto Y, Sugiura H, Hara Y, Wu X, Kobayashi S, Iwase H (2003) Quantitative determination, by real-time reverse transcription polymerase chain reaction, of aromatase mRNA in invasive ductal carcinoma of the breast. Breast Cancer Res 5:R250–R256. https://doi.org/10.1186/bcr657
Salhab M, Reed MJ, Al Sarakbi W, Jiang WG, Mokbel K (2006) The role of aromatase and 17-beta-hydroxysteroid dehydrogenase type 1 mRNA expression in predicting the clinical outcome of human breast cancer. Breast Cancer Res Treat 99:155–162. https://doi.org/10.1007/s10549-006-9198-8
Lu J, Li H, Cao D, Di G, Wu J, Sheng K, Han Q, Shen Z, Shao Z (2007) Clinical significance of aromatase protein expression in axillary node negative breast cancer. J Cancer Res Clin Oncol 133:401–409. https://doi.org/10.1007/s00432-006-0186-5
Licznerska BE, Wegman PP, Nordenskjold B, Wingren S (2008) In situ levels of oestrogen producing enzymes and its prognostic significance in postmenopausal breast cancer patients. Breast Cancer Res Treat 112:15–23. https://doi.org/10.1007/s10549-007-9819-x
Bollet MA, Savignoni A, De Koning L, Tran-Perennou C, Barbaroux C, Degeorges A, Sigal-Zafrani B, Almouzni G, Cottu P, Salmon R, Servant N, Fourquet A, de Cremoux P (2009) Tumor aromatase expression as a prognostic factor for local control in young breast cancer patients after breast-conserving treatment. Breast Cancer Res 11:R54. https://doi.org/10.1186/bcr2343
Girault I, Lerebours F, Tozlu S, Spyratos F, Tubiana-Hulin M, Lidereau R, Bieche I (2002) Real-time reverse transcription PCR assay of CYP19 expression: application to a well-defined series of post-menopausal breast carcinomas. J Steroid Biochem Mol Biol 82:323–332
Haiman CA, Dossus L, Setiawan VW, Stram DO, Dunning AM, Thomas G, Thun MJ, Albanes D, Altshuler D, Ardanaz E, Boeing H, Buring J, Burtt N, Calle EE, Chanock S, Clavel-Chapelon F, Colditz GA, Cox DG, Feigelson HS, Hankinson SE, Hayes RB, Henderson BE, Hirschhorn JN, Hoover R, Hunter DJ, Kaaks R, Kolonel LN, Le Marchand L, Lenner P, Lund E, Panico S, Peeters PH, Pike MC, Riboli E, Tjonneland A, Travis R, Trichopoulos D, Wacholder S, Ziegler RG (2007) Genetic variation at the CYP19A1 locus predicts circulating estrogen levels but not breast cancer risk in postmenopausal women. Cancer Res 67:1893–1897. https://doi.org/10.1158/0008-5472.CAN-06-4123
Prescott J, Thompson DJ, Kraft P, Chanock SJ, Audley T, Brown J, Leyland J, Folkerd E, Doody D, Hankinson SE, Hunter DJ, Jacobs KB, Dowsett M, Cox DG, Easton DF, De Vivo I (2012) Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS One 7:e37815. https://doi.org/10.1371/journal.pone.0037815
Flote VG, Furberg AS, McTiernan A, Frydenberg H, Ursin G, Iversen A, Lofteroed T, Ellison PT, Wist EA, Egeland T, Wilsgaard T, Makar KW, Chang-Claude J, Thune I (2014) Gene variations in oestrogen pathways, CYP19A1, daily 17beta-estradiol and mammographic density phenotypes in premenopausal women. Breast Cancer Res 16:499. https://doi.org/10.1186/s13058-014-0499-2
Thompson DJ, O'Mara TA, Glubb DM, Painter JN, Cheng T, Folkerd E, Doody D, Dennis J, Webb PM, Gorman M, Martin L, Hodgson S, Michailidou K, Tyrer JP, Maranian MJ, Hall P, Czene K, Darabi H, Li J, Fasching PA, Hein A, Beckmann MW, Ekici AB, Dork T, Hillemanns P, Durst M, Runnebaum I, Zhao H, Depreeuw J, Schrauwen S, Amant F, Goode EL, Fridley BL, Dowdy SC, Winham SJ, Salvesen HB, Trovik J, Njolstad TS, Werner HM, Ashton K, Proietto T, Otton G, Carvajal-Carmona L, Tham E, Liu T, Mints M, Scott RJ, McEvoy M, Attia J, Holliday EG, Montgomery GW, Martin NG, Nyholt DR, Henders AK, Hopper JL, Traficante N, Ruebner M, Swerdlow AJ, Burwinkel B, Brenner H, Meindl A, Brauch H, Lindblom A, Lambrechts D, Chang-Claude J, Couch FJ, Giles GG, Kristensen VN, Cox A, Bolla MK, Wang Q, Bojesen SE, Shah M, Luben R, Khaw KT, Pharoah PD, Dunning AM, Tomlinson I, Dowsett M, Easton DF, Spurdle AB (2016) CYP19A1 fine-mapping and Mendelian randomization: estradiol is causal for endometrial cancer. Endocr Relat Cancer 23:77–91. https://doi.org/10.1530/ERC-15-0386
Varghese JS, Smith PL, Folkerd E, Brown J, Leyland J, Audley T, Warren RM, Dowsett M, Easton DF, Thompson DJ (2012) The heritability of mammographic breast density and circulating sex-hormone levels: two independent breast cancer risk factors. Cancer Epidemiol Biomark Prev 21:2167–2175. https://doi.org/10.1158/1055-9965.EPI-12-0789
Kristensen VN, Harada N, Yoshimura N, Haraldsen E, Lonning PE, Erikstein B, Karesen R, Kristensen T, Borresen-Dale AL (2000) Genetic variants of CYP19 (aromatase) and breast cancer risk. Oncogene 19:1329–1333. https://doi.org/10.1038/sj.onc.1203425
Haiman CA, Hankinson SE, Spiegelman D, Brown M, Hunter DJ (2002) No association between a single nucleotide polymorphism in CYP19 and breast cancer risk. Cancer Epidemiol Biomark Prev 11:215–216
Chen C, Sakoda LC, Doherty JA, Loomis MM, Fish S, Ray RM, Lin MG, Fan W, Zhao LP, Gao DL, Stalsberg H, Feng Z, Thomas DB (2008) Genetic variation in CYP19A1 and risk of breast cancer and fibrocystic breast conditions among women in Shanghai, China. Cancer Epidemiol Biomark Prev 17:3457–3466. https://doi.org/10.1158/1055-9965.EPI-08-0517
Zhang L, Gu L, Qian B, Hao X, Zhang W, Wei Q, Chen K (2009) Association of genetic polymorphisms of ER-alpha and the estradiol-synthesizing enzyme genes CYP17 and CYP19 with breast cancer risk in Chinese women. Breast Cancer Res Treat 114:327–338. https://doi.org/10.1007/s10549-008-9998-0
Clendenen T, Zeleniuch-Jacquotte A, Wirgin I, Koenig KL, Afanasyeva Y, Lundin E, Arslan AA, Axelsson T, Forsti A, Hallmans G, Hemminki K, Lenner P, Roy N, Shore RE, Chen Y (2013) Genetic variants in hormone-related genes and risk of breast cancer. PLoS One 8:e69367. https://doi.org/10.1371/journal.pone.0069367
Pineda B, Garcia-Perez MA, Cano A, Lluch A, Eroles P (2013) Associations between aromatase CYP19 rs10046 polymorphism and breast cancer risk: from a case-control to a meta-analysis of 20,098 subjects. PLoS One 8:e53902. https://doi.org/10.1371/journal.pone.0053902
Zins K, Mogg M, Schneeberger C, Abraham D, Schreiber M (2014) Analysis of the rs10046 polymorphism of aromatase (CYP19) in premenopausal onset of human breast cancer. Int J Mol Sci 15:712–724. https://doi.org/10.3390/ijms15010712
Lukanova A, Lundin E, Micheli A, Arslan A, Ferrari P, Rinaldi S, Krogh V, Lenner P, Shore RE, Biessy C, Muti P, Riboli E, Koenig KL, Levitz M, Stattin P, Berrino F, Hallmans G, Kaaks R, Toniolo P, Zeleniuch-Jacquotte A (2004) Circulating levels of sex steroid hormones and risk of endometrial cancer in postmenopausal women. Int J Cancer 108:425–432. https://doi.org/10.1002/ijc.11529
Fasching PA, Loehberg CR, Strissel PL, Lux MP, Bani MR, Schrauder M, Geiler S, Ringleff K, Oeser S, Weihbrecht S, Schulz-Wendtland R, Hartmann A, Beckmann MW, Strick R (2008) Single nucleotide polymorphisms of the aromatase gene (CYP19A1), HER2/neu status, and prognosis in breast cancer patients. Breast Cancer Res Treat 112:89–98. https://doi.org/10.1007/s10549-007-9822-2
Long JR, Kataoka N, Shu XO, Wen W, Gao YT, Cai Q, Zheng W (2006) Genetic polymorphisms of the CYP19A1 gene and breast cancer survival. Cancer Epidemiol Biomark Prev 15:2115–2122. https://doi.org/10.1158/1055-9965.EPI-06-0464
Kuo SH, Yang SY, Lien HC, Lo C, Lin CH, Lu YS, Cheng AL, Chang KJ, Huang CS (2013) CYP19 genetic polymorphism haplotype AASA is associated with a poor prognosis in premenopausal women with lymph node-negative, hormone receptor-positive breast cancer. Biomed Res Int 2013:562197. https://doi.org/10.1155/2013/562197
Udler MS, Azzato EM, Healey CS, Ahmed S, Pooley KA, Greenberg D, Shah M, Teschendorff AE, Caldas C, Dunning AM, Ostrander EA, Caporaso NE, Easton D, Pharoah PD (2009) Common germline polymorphisms in COMT, CYP19A1, ESR1, PGR, SULT1E1 and STS and survival after a diagnosis of breast cancer. Int J Cancer 125:2687–2696. https://doi.org/10.1002/ijc.24678
Leyland-Jones B, Gray KP, Abramovitz M, Bouzyk M, Young B, Long B, Kammler R, Dell'Orto P, Biasi MO, Thurlimann B, Lyng MB, Ditzel HJ, Harvey VJ, Neven P, Treilleux I, Rasmussen BB, Maibach R, Price KN, Coates AS, Goldhirsch A, Pagani O, Viale G, Rae JM, Regan MM (2015) CYP19A1 polymorphisms and clinical outcomes in postmenopausal women with hormone receptor-positive breast cancer in the BIG 1-98 trial. Breast Cancer Res Treat 151:373–384. https://doi.org/10.1007/s10549-015-3378-3
Proestling K, Hebar A, Pruckner N, Marton E, Vinatzer U, Schreiber M (2012) The pro allele of the p53 codon 72 polymorphism is associated with decreased intratumoral expression of BAX and p21, and increased breast cancer risk. PLoS One 7:e47325. https://doi.org/10.1371/journal.pone.0047325
Vinatzer U, Dampier B, Streubel B, Pacher M, Seewald MJ, Stratowa C, Kaserer K, Schreiber M (2005) Expression of HER2 and the coamplified genes GRB7 and MLN64 in human breast cancer: quantitative real-time reverse transcription-PCR as a diagnostic alternative to immunohistochemistry and fluorescence in situ hybridization. Clin Cancer Res 11:8348–8357. https://doi.org/10.1158/1078-0432.CCR-05-0841
Stampfer MR, Bartley JC (1988) Human mammary epithelial cells in culture: differentiation and transformation. Cancer Treat Res 40:1–24
Pacher M, Seewald MJ, Mikula M, Oehler S, Mogg M, Vinatzer U, Eger A, Schweifer N, Varecka R, Sommergruber W, Mikulits W, Schreiber M (2007) Impact of constitutive IGF1/IGF2 stimulation on the transcriptional program of human breast cancer cells. Carcinogenesis 28:49–59. https://doi.org/10.1093/carcin/bgl091
Friesenhengst A, Pribitzer-Winner T, Schreiber M (2014) Association of the G473A polymorphism and expression of lysyl oxidase with breast cancer risk and survival in European women: a hospital-based case-control study. PLoS One 9:e105579. https://doi.org/10.1371/journal.pone.0105579
R Development Core Team (2009) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Harrington DP, Fleming TR (1982) A class of rank test procedures for censored survival-data. Biometrika 69:553–566
Lacroix M, Leclercq G (2004) Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat 83:249–289. https://doi.org/10.1023/B:BREA.0000014042.54925.cc
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515–527. https://doi.org/10.1016/j.ccr.2006.10.008
Miller WR, Anderson TJ, Jack WJ (1990) Relationship between tumour aromatase activity, tumour characteristics and response to therapy. J Steroid Biochem Mol Biol 37:1055–1059
Sasano H, Nagura H, Harada N, Goukon Y, Kimura M (1994) Immunolocalization of aromatase and other steroidogenic enzymes in human breast disorders. Hum Pathol 25:530–535
Brodie A, Lu Q, Nakamura J (1997) Aromatase in the normal breast and breast cancer. J Steroid Biochem Mol Biol 61:281–286
Ellis MJ, Miller WR, Tao Y, Evans DB, Chaudri Ross HA, Miki Y, Suzuki T, Sasano H (2009) Aromatase expression and outcomes in the P024 neoadjuvant endocrine therapy trial. Breast Cancer Res Treat 116:371–378. https://doi.org/10.1007/s10549-008-0161-8
Nebert DW, Russell DW (2002) Clinical importance of the cytochromes P450. Lancet 360:1155–1162. https://doi.org/10.1016/S0140-6736(02)11203-7
Blackburn HL, Ellsworth DL, Shriver CD, Ellsworth RE (2015) Role of cytochrome P450 genes in breast cancer etiology and treatment: effects on estrogen biosynthesis, metabolism, and response to endocrine therapy. Cancer Causes Control 26:319–332. https://doi.org/10.1007/s10552-014-0519-7
Germain D (2011) Estrogen carcinogenesis in breast cancer. Endocrinol Metab Clin N Am 40(473–484):vii. https://doi.org/10.1016/j.ecl.2011.05.009
Yoshimura N, Harada N, Bukholm I, Karesen R, Borresen-Dale AL, Kristensen VN (2004) Intratumoural mRNA expression of genes from the oestradiol metabolic pathway and clinical and histopathological parameters of breast cancer. Breast Cancer Res 6:R46–R55. https://doi.org/10.1186/bcr746
Miller WR, Hawkins RA, Forrest AP (1982) Significance of aromatase activity in human breast cancer. Cancer Res 42:3365s–3368s
Lykkesfeldt AE, Henriksen KL, Rasmussen BB, Sasano H, Evans DB, Moller S, Ejlertsen B, Mouridsen HT (2009) In situ aromatase expression in primary tumor is associated with estrogen receptor expression but is not predictive of response to endocrine therapy in advanced breast cancer. BMC Cancer 9:185. https://doi.org/10.1186/1471-2407-9-185
Silva MC, Rowlands MG, Dowsett M, Gusterson B, McKinna JA, Fryatt I, Coombes RC (1989) Intratumoral aromatase as a prognostic factor in human breast carcinoma. Cancer Res 49:2588–2591
Honma N, Takubo K, Sawabe M, Arai T, Akiyama F, Sakamoto G, Utsumi T, Yoshimura N, Harada N (2006) Estrogen-metabolizing enzymes in breast cancers from women over the age of 80 years. J Clin Endocrinol Metab 91:607–613. https://doi.org/10.1210/jc.2005-1967
Hudelist G, Wulfing P, Kersting C, Burger H, Mattsson B, Czerwenka K, Kubista E, Gschwantler-Kaulich D, Fink-Retter A, Singer CF (2008) Expression of aromatase and estrogen sulfotransferase in preinvasive and invasive breast cancer. J Cancer Res Clin Oncol 134:67–73. https://doi.org/10.1007/s00432-007-0249-2
Sasano H, Frost AR, Saitoh R, Harada N, Poutanen M, Vihko R, Bulun SE, Silverberg SG, Nagura H (1996) Aromatase and 17 beta-hydroxysteroid dehydrogenase type 1 in human breast carcinoma. J Clin Endocrinol Metab 81:4042–4046. https://doi.org/10.1210/jcem.81.11.8923858
Straume AH, Knappskog S, Lonning PE (2012) Effect of CYP19 rs6493497 and rs7176005 haplotype status on in vivo aromatase transcription, plasma and tissue estrogen levels in postmenopausal women. J Steroid Biochem Mol Biol 128:69–75. https://doi.org/10.1016/j.jsbmb.2011.08.015
Acknowledgements
Open access funding provided by Medical University of Vienna. The authors thank Martha R. Stampfer, Lawrence Berkeley National Laboratory, Berkeley, California, for kindly providing HMEC cells.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was approved and is annually reviewed by the Institutional Review Board (IRB, “Ethikkommission”) of the Medical University of Vienna, Austria (MUV; protocol 141/2002). Written informed consent was obtained from patients recruited after the onset of the study. For those patients who had undergone surgery before the onset of the study, a waiver of specific informed consent was approved by the IRB.
Financial Information
This work was supported by the Austrian Ministry of Science, Research, and Economy (Austrian Genome Research Program GEN-AU) (http://www.gen-au.at/) and the Anniversary Fund of the “Oesterreichische Nationalbank” (ID# 12994) (http://www.oenb.at/en/About-Us/Research-Promotion/The-OeNB-Anniversary-Fund.html.) The funders had no role in study design, the collection, analysis and interpretation of data, decision to submit for publication, or preparation of the manuscript.
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
ESM 1
(PDF 1675 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Friesenhengst, A., Pribitzer-Winner, T., Miedl, H. et al. Elevated Aromatase (CYP19A1) Expression Is Associated with a Poor Survival of Patients with Estrogen Receptor Positive Breast Cancer. HORM CANC 9, 128–138 (2018). https://doi.org/10.1007/s12672-017-0317-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12672-017-0317-2