Abstract
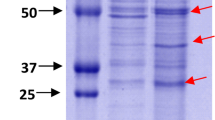
Bacterial intracellular proteinases play a relevant role in coordinating various cellular processes. The genome of the uropathogenic Klebsiella oxytoca NK-1 encodes the gene prtKO, homologous to grimelysin of Serratia grimesii. BLAST analysis showed that a lot of bacteria encode grimelysin-like proteinases, known to be mainly distributed in Enterobacterales. Phylogenetic analysis classified these proteins into 10 clusters. PrtKO was predicted to contain conservative motifs typical for grimelysin using bioinformatic tools. In addition, the structure of metalloproteinase-containing genomic loci is conservative among enterobacteria and consists of the genes involved in global physiological processes. Maximal total intracellular proteolytic activity of K. oxytoca NK-1 is observed during the late stationary phase of growth in LB medium at 37 °C. Limited proteolysis of actin indicates the presence of a grimelysin-like proteinase in bacterial cells during the late stationary growth phase. Hence, we present a first report of a new grimelysin-like intracellular metalloproteinase in K. oxytoca.






Similar content being viewed by others
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Code Availability
Not applicable.
References
Adeolu, M., Alnajar, S., Naushad, S., & Gupta, R. S. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. International Journal of Systematic and Evolutionary Microbiology, 66, 5575–5599. https://doi.org/10.1099/ijsem.0.001485
Bozhokina, E., Khaitlina, S., & Adam, T. (2008). Grimelysin, a novel metalloprotease from Serratia grimesii, is similar to ECP32. Biochemical and Biophysical Research Communications, 367, 888–892. https://doi.org/10.1016/j.bbrc.2008.01.003
Bozhokina, E. S., Tsaplina, O. A., Efremova, T. N., et al. (2011). Bacterial invasion of eukaryotic cells can be mediated by actin-hydrolysing metalloproteases grimelysin and protealysin. Cell Biology International, 35, 111–118. https://doi.org/10.1042/CBI20100314
Bozhokina, E., Kever, L., & Khaitlina, S. (2020). The Serratia grimesii outer membrane vesicles-associated grimelysin triggers bacterial invasion of eukaryotic cells. Cell Biology International, 44, 2275–2283. https://doi.org/10.1002/cbin.11435
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Breidenstein, E. B., Janot, L., Strehmel, J., et al. (2012). The Lon protease is essential for full virulence in Pseudomonas aeruginosa. PLoS ONE, 7, e49123. https://doi.org/10.1371/journal.pone.0049123
Chiavolini, D., Memmi, G., Maggi, T., Iannelli, F., Pozzi, G., & Oggioni, M. R. (2003). The three extra-cellular zinc metalloproteinases of Streptococcus pneumoniae have a different impact on virulence in mice. BMC Microbiology, 3, 14. https://doi.org/10.1186/1471-2180-3-14
Chukhontseva, K. N., Salnikov, V. V., Morenkov, O. S., Kostrov, S. V., & Demidyuk, I. V. (2019). Protealysin is not secreted constitutively. Protein and Peptide Letters, 26, 221–226. https://doi.org/10.2174/0929866526666181212114907
Demidyuk, I. V., Kalashnikov, A. E., Gromova, T. Y., et al. (2006). Cloning, sequencing, expression, and characterization of protealysin, a novel neutral proteinase from Serratia proteamaculans representing a new group of thermolysin-like proteases with short N-terminal region of precursor. Protein Expression and Purification, 47, 551–561.
Demidyuk, I., Gasanov, E., Safina, D., & Kostrov, S. (2008). Structural organization of precursors of thermolysin-like proteinases. Protein Journal, 27, 343–354. https://doi.org/10.1007/s10930-008-9143-2
Demidyuk, I., Gromova, T., Polyakov, M., et al. (2010). Crystal structure of the protealysin precursor: Insights into propeptide function. Journal of Biological Chemistry, 285, 2003–2013. https://doi.org/10.1074/jbc.M109.015396
Eckhard, U., Huesgen, P. F., Brandstetter, H., & Overall, C. M. (2014). Proteomic protease specificity profiling of clostridial collagenases reveals their intrinsic nature as dedicated degraders of collagen. Journal of Proteomics, 100, 102–114. https://doi.org/10.1016/j.jprot.2013.10.004
Giliazeva, A. G., Shagimardanova, E. I., Shigapova, L. H., et al. (2019). Draft genome sequence and analysis of Klebsiella oxytoca strain NK-1 isolated from ureteral stent. Data in Brief, 24, 103853. https://doi.org/10.1016/j.dib.2019.103853
Gong, Y., Xu, W., Cui, Y., et al. (2011). Immunization with a ZmpB-based protein vaccine could protect against pneumococcal diseases in mice. Infection and Immunity, 79, 867–878. https://doi.org/10.1128/IAI.00717-10
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Hering, N. A., Fromm, A., Bücker, R. et al. (2019). Tilivalline- and tilimycin-independent effects of Klebsiella oxytoca on tight junction-mediated intestinal barrier impairment.https://doi.org/10.3390/ijms20225595
Khaitlina, SYu., Smirnova, T. D., & Usmanova, A. M. (1988). Limited proteolysis of actin by a specific bacterial protease. FEBS Letters, 228, 172–174.
Khaitlina, S., Bozhokina, E., Tsaplina, O., & Efremova, T. (2020). Bacterial actin-specific endoproteases grimelysin and protealysin as virulence factors contributing to the invasive activities of Serratia. International Journal of Molecular Sciences, 21, 4025. https://doi.org/10.3390/ijms21114025
Konovalova, A., Søgaard-Andersen, L., & Kroos, L. (2014). Regulated proteolysis in bacterial development. FEMS Microbiology Reviews, 38, 493–522. https://doi.org/10.1111/1574-6976.12050
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. https://doi.org/10.1093/molbev/msw054
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Lloubès, R., Goemaere, E., Zhang, X., Cascales, E., & Duché, D. (2012). Energetics of colicin import revealed by genetic cross-complementation between the Tol and Ton systems. Biochemical Society Transactions, 40, 1480–1485. https://doi.org/10.1042/BST20120181
Malekjamshidi, M. R., Zandi, H., & Eftekhar, F. (2020). Prevalence of extended-spectrum β-lactamase and integron gene carriage in multidrug-resistant Klebsiella species isolated from outpatients in Yazd, Iran. Iranian Journal of Medical Sciences, 45, 23–31. https://doi.org/10.30476/IJMS.2019.45334
Marshall, N. C., Finlay, B. B., & Overall, C. M. (2017). Sharpening host defenses during infection: Proteases cut to the chase. Molecular and Cellular Proteomics, 16, S161–S171. https://doi.org/10.1074/mcp.O116.066456
Odoki, M., Almustapha Aliero, A., Tibyangye, J., et al. (2019). Prevalence of bacterial urinary tract infections and associated factors among patients attending hospitals in Bushenyi District, Uganda. International Journal of Microbiology, 2019, 4246780. https://doi.org/10.1155/2019/4246780
Okonechnikov, K., Golosova, O., Fursov, M., & UGENE team. (2012). Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics, 28, 1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Pan, S., Malik, I. T., Thomy, D., Henrichfreise, B., & Sass, P. (2019). The functional ClpXP protease of Chlamydia trachomatis requires distinct clpP genes from separate genetic loci. Science and Reports, 9, 14129. https://doi.org/10.1038/s41598-019-50505-5
Petersen, L. M., & Tisa, L. S. (2013). Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Canadian Journal of Microbiology, 59, 627–640. https://doi.org/10.1139/cjm-2013-0343
Sekowska, A., Gospodarek, E., Janickca, G., Jachna-Sawicka, K., & Sawicki, M. (2006). Hydrolytic and haemolytic activity of Klebsiella pneumoniae and Klebsiella oxytoca. Medycyna Doswiadczalna I Mikrobiologia, 58(2), 135–141.
Subramanian, K., Henriques-Normark, B., & Normark, S. (2019). Emerging concepts in the pathogenesis of the Streptococcus pneumoniae: From nasopharyngeal colonizer to intracellular pathogen. Cellular Microbiology, 21, e13077. https://doi.org/10.1111/cmi.13077
Sullivan, M. J., Petty, N. K., & Beatson, S. A. (2011). Easyfig: A genome comparison visualizer. Bioinformatics, 27, 1009–1010. https://doi.org/10.1093/bioinformatics/btr039
Tian, Z., Cheng, S., Xia, B., et al. (2019). Pseudomonas aeruginosa ExsA regulates a metalloprotease, ImpA, that inhibits phagocytosis of macrophages. Infection and Immunity. https://doi.org/10.1128/IAI.00695-19
Tsaplina, O. A., Efremova, T. N., Kever, L. V., et al. (2009). Probing for actinase activity of protealysin. Biochemistry (Moscow), 74, 648–654. https://doi.org/10.1134/s0006297909060091
Tsaplina, O., Efremova, T., Demidyuk, I., & Khaitlina, S. (2012). Filamentous actin is a substrate for protealysin, a metalloprotease of invasive Serratia proteamaculans. FEBS Journal, 279, 264–274. https://doi.org/10.1111/j.1742-4658.2011.08420.x
Wyres, K. L., Lam, M. M., & Holt, K. E. (2020). Population genomics of Klebsiella pneumoniae. Nature Reviews Microbiology, 18, 344–359. https://doi.org/10.1038/s41579-019-0315-1
Wysocki, A. B., Bhalla-Regev, S. K., Tierno, P. M., Jr., et al. (2013). Proteolytic activity by multiple bacterial species isolated from chronic venous leg ulcers degrades matrix substrates. Biological Research for Nursing, 15, 407–415. https://doi.org/10.1177/1099800412464683
Acknowledgements
This work was performed in accordance with the Russian Government Program of Competitive Growth of Kazan Federal University. We express our sincere gratitude to Yaw Akosah for his thoughtful comments and efforts toward improving our manuscript.
Funding
The study was funded by Russian Foundation for Basic Research (Project No. 18–34-00837).
Author information
Authors and Affiliations
Contributions
A. Mardanova and M. Sharipova conceived and designed the experiments and analyzed the data. Material preparation and data collection were performed by P. Misheeva. A. Giliazeva performed the experiments, analyzed and interpreted the data, and wrote the paper. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Research Involving Human and Animals Statement
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Informed Consent
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the Topical Collection: Molecular, Precision and Regenerative Medicine - 2021
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Giliazeva, A.G., Misheeva, P.S., Sharipova, M.R. et al. Bioinformatic Analysis of a Grimelysin-like Protease in the Klebsiella oxytoca Strain NK-1. BioNanoSci. 12, 160–169 (2022). https://doi.org/10.1007/s12668-021-00924-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12668-021-00924-4