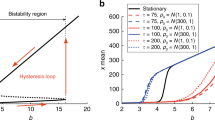
Abstract
Multicellular organisms exhibit a high degree of structural organization with specific cell types always occurring in characteristic locations. The conventional framework for describing the emergence of such consistent spatial patterns is provided by Wolpert’s “French flag” paradigm. According to this view, intra-cellular genetic regulatory mechanisms use positional information provided by morphogen concentration gradients to differentially express distinct fates, resulting in a characteristic pattern of differentiated cells. However, recent experiments have shown that suppression of inter-cellular interactions can alter these spatial patterns, suggesting that cell fates are not exclusively determined by the regulation of gene expression by local morphogen concentration. Using an explicit model where adjacent cells communicate by Notch signaling, we provide a mechanistic description of how contact-mediated interactions allow information from the cellular environment to be incorporated into cell fate decisions. Viewing cellular differentiation in terms of trajectories along an epigenetic landscape (as first enunciated by Waddington), our results suggest that the contours of the landscape are molded differently in a cell position-dependent manner, not only by the global signal provided by the morphogen but also by the local environment via cell–cell interactions. We show that our results are robust with respect to different choices of coupling between the inter-cellular signaling apparatus and the intra-cellular gene regulatory dynamics. Indeed, we show that the broad features can be observed even in abstract spin models. Our work reconciles interaction-mediated self-organized pattern formation with boundary-organized mechanisms involving signals that break symmetry.






Similar content being viewed by others
References
L Wolpert The Triumph of the Embryo (Oxford: Oxford University Press) (1991)
L Wolpert and C Tickle Developmental Biology (Oxford: Oxford University Press) (2011)
S Gilbert Developmental Biology (MA.: Sinauer, Sunderland) (2013)
C H Waddington Organisers and Genes (Cambridge: Cambridge University Press) (1940)
A M Turing. Philos. Trans. R. Soc. London B 237 37 (1952).
M C Cross and P C Hohenberg.Rev. Mod. Phys. 65 851 (1993).
A J Koch and H Meinhardt. Rev. Mod. Phys. 66 1481 (1994).
P Ball The Self-made Tapestry: Pattern Formation in Nature (Oxford University Press, Oxford, 1999). https://doi.org/10.1119/1.880339
H T Zhang and T Hiiragi. Annu. Rev. Cell Dev. Biol. 34 405 (2018).
F Crick. 225 420 (1970).
W Driever and C Nüsslein-Volhard. Cell 54 95 (1988).
A A Teleman, M Strigini and S M Cohen. Cell 105 559 (2001).
A D Lander, Q Nie and F Y M Wan. Dev. Cell 2 785 (2002).
T Bollenbach, K Kruse, P Pantazis, M González-Gaitán and F Jülicher. Phys. Rev. Lett. 94 018103 (2005).
J L England and J Cardy. Phys. Rev. Lett. 94 078101 (2005).
G Hornung, B Berkowitz and N Barkai.Phys. Rev. E 72 041916 (2005).
A D Lander. Cell 128 245 (2007).
D Ben-Zvi and N Barkai. Proc. Natl. Acad. Sci. USA 107 6924 (2010).
S B Yuste, E Abad and K Lindenberg. Phys. Rev. E 82 061123 (2010).
C B Muratov, P V Gordon and S Y Shvartsman. Phys. Rev. E 84 041916 (2011).
A D Lander. Cell 144 955 (2011).
C Kuyyamudi, S N Menon, F Casares and S Sinha.Phys. Rev. E 104 L052401 (2021).
L Wolpert. J. Theor. Biol. 25 1 (1969).
L Wolpert. Development 107 3 (1989).
J Sharpe. Development, 146, dev185967 (2019).
J B Gurdon and P-Y Bourillot. Nature (Lond.) 413, 797 (2001).
H L Ashe and J Briscoe. Developement 133 385 (2006).
K W Rogers and A F Schier. Annu. Rev. Cell Dev. Biol. 27 377 (2011).
J H Kong, L Yang, E Dessaud, K Chuang, D M Moore, R Rohatgi, J Briscoe and B G. Dev. Cell 33 373 (2015).
H Meinhardt Models of Biological Pattern Formation (London: Academic Press) (1982)
S Werner, T Stückemann, M B Amigo, J C Rink, F Jülicher and B M Friedrich. Phys. Rev. Lett. 114 138101 (2015).
S Artavanis-Tsakonas, M D Rand and R J Lake. Science 284 770 (1999).
R Kopan and M X G Ilagan. Cell 137 216 (2009).
D Sprinzak, A Lakhanpal, L LeBon, J Garcia-Ojalvo and M B Elowitz. PLoS Comput. Biol. 7 e1002069 (2011).
T Erdmann, M Howard and P R Ten Wolde. Phys. Rev. Lett. 103 258101 (2009).
A D Lander. Science 339 923 (2013).
E Dessaud, A P McMahon and J Briscoe. Development 135 2489 (2008).
E Dessaud, V Ribes, N Balaskas, L L Yang, A Pierani, A Kicheva, B G Novitch, J Briscoe and N Sasai. PLoS Biol. 8 e1000382 (2010).
N Balaskas, A Ribeiro, J Panovska, E Dessaud, N Sasai, K M Page, J Briscoe and V Ribes. Cell 148 273 (2012).
R Phillips, J Kondev, J Theriot, H G Garcia and N Orme Physical Biology of the Cell (New York, NY: Garland Science) (2013)
H Shimojo, T Ohtsuka and R Kageyama .Front. Neurosci. 5 78 (2011).
L J Manderfield, F A High, K A Engleka, F Liu, L Li, S Rentschler and J A Epstein. Circulation 125 314 (2012).
M Boareto, M K Jolly, M Lu, J N Onuchic, C Clementi and E Ben-Jacob. Proc. Natl. Acad. Sci. USA 112 E402 (2015).
C Kuyyamudi, S N Menon and S Sinha. Phys. Rev. E 103 062409 (2021).
J R Collier, N A Monk, P K Maini and J H Lewis. J. Theor. Biol. 183 429 (1996).
D Sprinzak, A Lakhanpal, L LeBon, L A Santat, M E Fontes, G A Anderson, J Garcia-Ojalvo and M B Elowitz. Nature 465 86 (2010).
C Kuyyamudi, S N Menon and S Sinha. (2021).
M B Miller and B L Bassler. Annu. Rev. Microbiol. 55, 165 (2001).
C M Waters and B L Bassler. Annu. Rev. Cell Dev. Bi. 21 319 (2005).
J Blanchard. Phys. Rev. 11, 81 (1918).
F S Werblin. Sci. Am. 228 70 (1973).
J B Green and J Sharpe. Development 142 1203 (2015).
C Kuyyamudi, S N Menon and S Sinha. Phys. Biol. 19 016001 (2022).
C H Waddington The Strategy of the Genes (London: George Allen & Unwin) (1957)
J E Ferrell. Curr. Biol. 22 R458 (2012).
P Wang, C Song, H Zhang, Z Wu, X J Tian and J Xing. Interface Focus 4 20130068 (2014).
M Mojtahedi, A Skupin, J Zhou, I G Casta\(\tilde{\text{n}}\)o, R Y Y Leong-Quong, H Chang, K Trachana, A Giuliani, and S Huang. PLoS Biol. 14, e2000640 (2016).
N Moris, C Pina and A M. Nat. Rev. Genet. 17 693 (2016).
Acknowledgements
We would like to thank Marcin Zagórski for helpful discussions. SNM has been supported by the IMSc Complex Systems Project (12th Plan), and the Center of Excellence in Complex Systems and Data Science, both funded by the Department of Atomic Energy, Government of India. The simulations required for this work were supported by IMSc High Performance Computing facility (hpc.imsc.res.in) [Nandadevi.]
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kuyyamudi, C., Menon, S.N. & Sinha, S. Flags, landscapes and signaling: contact-mediated inter-cellular interactions enable plasticity in fate determination driven by positional information. Indian J Phys 96, 2657–2666 (2022). https://doi.org/10.1007/s12648-022-02348-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12648-022-02348-6