Abstract
One of the major obstacles to malaria elimination in the world is the resistance in Plasmodium falciparum to most antimalarial drugs. This study aimed to estimate the prevalence of molecular markers of antimalarial drugs resistance in Côte d’Ivoire. Samples were collected from 2013 to 2016 from asymptomatic and symptomatic subjects in Abengourou, Abidjan, Grand Bassam, and San Pedro. A total of 704 participants aged between 1 year and 65 years (Mean age: 9 years ± 7.7) were enrolled. All the dried filter paper blood spots were genotyped by sequencing. Plasmodium falciparum kelch propeller domain 13 (pfk13) gene were analyzed for all the samples, while 344 samples were examined for Plasmodium falciparum multi-drug resistance 1 (pfmdr1). Overall, the success rate of molecular tests was 98.8% (340/344), 99.1% (341/344), and 94.3% (664/704) for pfmdr1 N86Y, pfmdr1 Y184F, and pfk13 genes respectively. Molecular analysis revealed twenty (5.9%; 20/340) and 219 (64.2%; 219/341) mutant alleles for pfmdr1 86Y and pfmdr1 184 F, respectively. Twenty-nine mutations in pfk13 gene (4.4%; 29/664) with 2.7% (18/664) of non-synonymous mutations was found. None of the mutations previously described in South East Asia (SEA) involved in P. falciparum resistance to artemisinin derivatives were observed in this study. According to year of collection, a decrease of the prevalence of pfk13 mutation (from 3.6 to 1.8%) and pfmdr1 N86Y mutation (from 8.5 to 4.5%) and an increase of mutant allele of pfmdr1 Y184F proportion (from 39.8 to 66.4%) were found. Comparing to previous studies in the country, this study showed an increase in lumefantrine tolerance of P. falciparum strains. This demonstrates the importance of establishing a strong system for molecular surveillance of malaria in Côte d’Ivoire.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malaria elimination remains threatened by the resistance of P. falciparum to most antimalarial drugs. To address this concern, most endemic countries have adopted artemisinin-based combination therapies (ACTs) as first-line treatment for uncomplicated malaria. Artesunate-amodiaquine (ASAQ) and artemether-lumefantrine (AL) were officially adopted as first-line treatment for uncomplicated malaria in Côte d’Ivoire since 2007 (Ministère de la Santé et de l’Hygiène Publique 2007). The efficacy of these combinations depends on the susceptibility of parasites to artemisinin derivatives and partners drugs. Artemisinin resistance has been confirmed in five countries in the Greater Mekong subregion, namely Cambodia, the Lao People’s Democratic Republic, Myanmar, Thailand, and Vietnam (Dondorp et al. 2011; Noedl et al. 2008; Plowe 2009). Recently, P. falciparum isolates with mutations in Plasmodium falciparum kelch propeller domain 13 (pfk13) gene linked to artemisinin resistance have been also reported in Rwanda and Tanzania (Uwimana et al. 2020; Bwire et al. 2020; Bergmann et al. 2021). Other sub-Saharan African countries including Côte d’Ivoire could similarly face the same phenomenon with a risk of increased malarial morbidity and mortality rates. The study of molecular markers is an early warning tool for drug resistance that can guide clinical efficacy studies on antimalarial drugs and malaria treatment guidelines (Ringwald et al. 2002). The molecular markers most commonly studied for antimalarial drug resistance are Plasmodium falciparum chloroquine resistance transporter (pfcrt), Plasmodium falciparum multi-drug resistance 1 (pfmdr1), and pfk13. Pfcrt is linked to chloroquine (CQ) and amodiaquine (AQ) resistance (Eyase et al. 2013). Pfmdr1 is involved in P. falciparum resistance to many antimalarial drugs, such as AQ and lumefantrine, which are used as partner drug for artemisinin derivatives in ASAQ and AL (Sisowath et al. 2005; Dokomajilar et al. 2006). Pfk13 gene is a molecular marker of artemisinin resistance (WHO 2017). In Côte d’Ivoire, few recent data are available on the allelic frequencies of these genes. In the past, pfcrt 76T mutant allele accounted for 47.6–100% in Abidjan (the economic capital) (Djaman et al. 2004; Bla et al. 2014) and in its border towns (Ako et al. 2012; Ouattara et al. 2011). Our preliminary study carried on symptomatic subjects showed a prevalence of pfcrt gene mutation of 16.6% (Konaté et al. 2018a, 2018b). Three points of mutations (N86Y, Y184F, and D1246Y) of pfmdr1 gene, with respectively frequencies of 47.5%, 75%, and 1.6% were reported in Côte d’Ivoire in 2006–2007 [unpublished data]. None of the mutations in pfk13 gene observed in South-East Asia (SEA) have been observed in Côte d’Ivoire (Kamau et al. 2014; Basco et al. 2017). Thus, this study aimed to update data by determining the prevalence of molecular markers of antimalarial drug resistance in Côte d’Ivoire.
Materials and methods
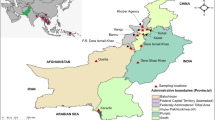
Study area
The study was conducted on samples collected at Abidjan, Grand-Bassam (coastal and forest zones), Abengourou (forest zone), and San Pedro (coastal and forest zones). Côte d’Ivoire experiences two wet seasons (from April to July and from October to November) and two dry seasons (from December to March and August to September). Malaria transmission in the country is perennial, with peaks occurring during wet seasons. According to the National Malaria Control Program (NMCP), malaria cases are predominantly caused by P. falciparum (95–99%), followed by P. malariae (3–4.2%) and P. ovale (0.3–0.7%) (Assi et al. 2013; Ministère de la Santé et de la Lutte contre le SIDA 2015).
Study design
Plasmodium falciparum infected samples were collected from asymptomatic and symptomatic individuals from 2013 to 2016. Molecular analysis was performed from May 2016 to November 2018 at the Malaria Research and Control Center located at the National Institute of Public Health (Abidjan, Côte d’Ivoire).
Samples from symptomatic subjects were collected through routine evaluation of antimalarial drugs at public health facilities in Abidjan (2013), and Abengourou and San Pedro (2016). The inclusion criteria of symptomatic patients were: an age between 6 months to 65 years, with uncomplicated malaria and a monospecific P. falciparum infection diagnosed by microscopy. Another requirement was that the patients must have maintained residence in the study area for at least 2 months.
Asymptomatic subjects were recruited at primary school in Abengourou, Grand-Bassam and San Pedro (2015–2016). The group largely comprised school children aged between 4 and 16 years. Any non-febrile (temporal temperature below 37.8 ºC) subject carrying the asexual form of Plasmodium without any clinical signs of malaria during the seven days before and after medical examination was considered asymptomatic.
Malaria confirmation
To confirm Plasmodium carriage and determine parasitemia, thick and thin blood films were performed. The density of P. falciparum in peripheral blood was determined by counting the number of asexual parasites for 200 white blood cells per µL; i.e., number of parasites × 8000/200 assumed a white blood cell mean of 8,000 cells per µL as recommended by WHO when the patient’s exact white blood cell count is not available (WHO 2010). A double-check reading was performed for all slides.
Dried filter paper blood spots collection
For molecular analysis and prior to any treatment (symptomatic subjects), approximately 50 µL of whole blood was collected from the subjects’ fingertips and blotted on sterile filter paper (N°3; Whatman International Ltd. Maidstone, UK). The blotted filter paper was then dried secluded from insects and dust for at least 24 h. Dried blood spots were then stored at room temperature, protected with a silica-gel desiccant, and archived for molecular analysis.
DNA extraction
Parasite DNA was extracted from dried blood spots using the QIAamp® DNA Blood Mini Kit (Qiagen, Hilden, Germany) and the Quick-DNA™ Universal Kit (Zymo Research, California, USA) according to the manufacturer’s instructions.
Genotyping of pfmdr1 and pfk13 genes
Pfk13 gene was analyzed on all the dried blood spots, while pfmdr1 polymorphism was studied on dried blood spots from Abidjan, Abengourou, and San Pedro. Nested PCR was done for all the samples. The manufacturer’s instructions were followed throughout the process. The first mix contained for each amplification 12.5 µL of the master mix named OneTaq® 2X Master Mix with Standard Buffer (New England Biolabs, Ipswich, MA, USA), 0.5 µL of each primer (10 µM), 6.5 µL of Nuclease free water, and 5 µL of DNA template. The second amplification used a final volume of 50 µL final volume sufficient for sequencing. The primer sequences are listed in Table 1. After the second amplification, electrophoresis enabled visualization of PCR products on a 1.5% agarose gel. Electrophoresis was performed at 100 mV for 45 min. All pfmdr1 and pfk13 PCR products that were successfully amplified and then verified by the visualization of a fragment after migration were selected for Sanger sequencing.
Sequencing and sequences analysis
The PCR products were sequenced according to the Sanger sequencing method by the company Genewiz (Takeley, United Kingdom). The sequences were analyzed by alignment with the BioEdit™ software using pf3D7_1343700 (NCBI) as a reference strain (Reference sequence accession number: XM_001350122 for pfk13 and S53996 for pfmdr1). Codons 86 and 184 for the pfmdr1 gene were analyzed while the entire sequence was analyzed for pfk13 gene.
Ethical considerations
The study was approved by the National Research Ethics Committee (Certificates numbers: N° 020/MSLS/CNER-dkn, N˚ 049/MSLS/CNER-dkn). The study was conducted in accordance with the principles of the Helsinki Declaration adopted by the 18th World Medical Assembly in 1964, its amendments (2000, amended in Tokyo in 2004), and the International Conference on Harmonization (ICH) recommendations. The design and execution of our study has been Good Clinical Practice compliant and all applicable regulatory requirements for clinical studies, as well as Côte d’Ivoire’s national laws and regulations have been met. Free and written informed consent was obtained from patients, parents, or legal guardians prior to the participant’s enrolment.
Statistical analysis
SPSS Statistics version 21 (IBM Corp., Armonk, NY, USA) was used for data analysis. Different parameters were compared using the Fisher’s exact test. The level of significance for the statistical tests was set at 0.05. The polymorphism of pfmdr1 and pfk13 genes was determined based on the presence or absence of the wild-type and mutant alleles at the loci.
Results
A total of 704 participants were enrolled in this study. The study population comprised 460 symptomatic subjects and 244 asymptomatic subjects, with a mean age of 9 years (SD = 7.7). The minimum and maximum ages recorded were 1 year and 65 years, respectively. The geometric mean parasite count was 4,953 (SD = 43,066; range, 16–240,000) trophozoites per microliter of blood. All samples were analyzed for pfk13 while 344 (all from symptomatic patients) were examined for pfmdr1. Overall, the success rates of molecular tests for pfmdr1 N86Y, pfmdr1 Y184F, and pfk13 were 98.8% (340/344), 99.1% (341/344), and 94.3% (664/704), respectively. Twenty (5.9%) 86Y and 219 (64.2%) 184 F mutant alleles of the pfmdr1 gene were found (Fig. 1). In addition, 29 mutants (4.4%; 29/664) of pfk13 gene were found, with 18 non-synonymous mutations (2.7%).
According to year of collection (from 2013 to 2016), a decrease of the prevalence of pfk13 mutation (from 3.6 to 1.8%) and pfmdr1 N86Y mutation (from 8.5 to 4.5%) was noticed while an increase of mutant allele of pfmdr1 Y184F proportion (from 39.8 to 66.4%) was found. The prevalence of pfk13 gene mutations was significantly associated with parasite density (p = 0.003; Table 2). A578S (three cases) and G665S (two cases) were the most frequently detected mutations. In asymptomatic participants, the prevalence of pfk13 gene mutations was 2.1% (5/243), of which 0.8% (2/243) were non-synonymous (Table 3). In the symptomatic group, the prevalence of pfk13 gene mutations was 5.7% (24/421), of which 2.1% (9/421) were non-synonymous. For Pfmdr1 gene, five non-common points of mutations were found apart from Pfmdr1 N86Y and Pfmdr1 Y184F: four cases of Pfmdr1 R133K and one (1) case of Pfmdr1 D156Y.
*Fisher’s exact test.
**only for symptomatic subjects
Discussion
Regular monitoring of P. falciparum resistance molecular markers is a key factor in the early detection of emerging resistance and achieving malaria elimination. In this study, pfmdr1 and pfk13 genes were studied for their implication in P. falciparum resistance to antimalarial drugs, particularly amodiaquine, lumefantrine, and artemisinin derivatives. The limitation of this study was the lack of data to investigate the temporal comparison in the same site to identify subnational trends in prevalence in different geographic areas.
A selection for pfmdr1 86 N wild-type alleles was observed as a result of using ASAQ and AL for many years. This selection is commonly associated with lumefantrine tolerance (Raman et al. 2019). Indeed, pfmdr1 polymorphism is usually studied because of its involvement in P. falciparum’s resistance to many antimalarial drugs, particularly those used as partner drugs of artemisinin derivatives in ASAQ and AL (amodiaquine and lumefantrine). Single-nucleotide polymorphisms (SNPs) at codons 86 (N → Y), 184 (Y → F), and 1246 (D → Y) of pfmdr1 are among the most observed. Monitoring of NFD and YYY haplotypes is useful for detecting an early decrease in susceptibility to lumefantrine and amodiaquine, respectively (Venkatesan et al. 2014). Usually, after the change from chloroquine to ACTs, an increase in parasites carrying pfmdr1 86 N was found, as reported in this study from 2013 to 2016 (Okell et al. 2018). In the same way, a previous study conducted in 2006 i.e. one year before the official adoption of ACTs as first-line treatment for uncomplicated malaria cases showed a higher value of the mdr186Y allele (47.5%) than ours (unpublished data). In addition, an upward trend of mdr184F mutant-allele prevalence is usually reported after the adoption of ACTs, as observed in the present study from 2013 to 2016 (Okell et al. 2018). The prevalence of this mutation in Abidjan in 2010 was 57% (Trebissou et al. 2014), which was lower than ours. In this study, we did not investigate the mdrD1246Y mutation. A previous study carried out in Côte d’Ivoire on this mutation reported a low incidence of mutant (1.6%) [unpublished data]. A scarcity of mutant mdr1246Y has also been found in Burkina Faso (Sondo et al. 2016). These results point to lumefantrine tolerance in strains circulating through the country, as found in many reports from sub-Saharan African areas that use AL combination (Raman et al. 2019; Sondo et al. 2016; Dama et al. 2017). The pfmdr1 gene results observed in this study could be due to frequent use of AL combination over AS + AQ due to patient complaints following the use of the latter (Azagoh-Kouadio et al. 2017). Moreover, ex vivo susceptibility tests of P. falciparum strains circulating in Côte d’Ivoire should confirm this result. In Mali, a reduced ex vivo susceptibility of P. falciparum was observed after AL treatment (Dama et al. 2017).
Mutations in pfk13 gene (C580Y, R561H, R539T, I543T, Y493H, M476I, N458Y, Y493H, and recently F446I) (Wang et al. 2018) are molecular markers of artemisinin resistance validated by in vitro and in vivo studies, while P441L, G449A, G538V, P553L, R561H, V568G, P574L, A578S, and A675V are candidate markers (WHO 2017). These validated mutations have been reported in Cambodia and other SEA countries and serve as tools for monitoring ACTs resistance (Ariey et al. 2014). These mutations are absent in this study as well as in a previous study conducted in 2017 in Abidjan (Basco et al. 2017). This result suggests that P. falciparum parasites circulating in Côte d’Ivoire are still sensitive to artemisinin. This is consistent with the findings of previous results from clinical trials at sentinel sites in the country that have shown adequate clinical and parasitological responses of up to 100% (Toure et al. 2014; Yavo et al. 2015; Konaté et al. 2018a). However, recent studies in Rwanda and Tanzania have showed the presence of R561H mutations (Uwimana et al. 2020; Bwire et al. 2020; Bergmann et al. 2021). In addition, phylogenetic analysis of Rwanda strains revealed the expansion of an indigenous R561H lineage and evidence for the de novo emergence of Pfkelch13-mediated artemisinin resistance in Rwanda (Uwimana et al. 2020, 2021). These data highlight the importance of surveying antimalarial molecular markers in Côte d’Ivoire. Additional mutations were also identified in this study. Indeed, A557S, a non-synonymous mutation already reported in Côte d’Ivoire (Kamau et al. 2014) and Congo by Taylor et al. (Taylor et al. 2015), was found in this study. In addition, the S522C mutation observed in this study has already been described in Africa, particularly West Africa (Togo), Central Africa (Central African Republic, Gabon, Democratic Republic of Congo), and East Africa (Kenya) (Menard et al. 2016). The A578S mutation, a candidate marker for artemisinin resistance (WHO 2017), has the highest prevalence in this study (0.5%). This mutation seems to be the most widespread worldwide. Indeed, it has been reported in a small proportion in Cambodia (Straimer et al. 2015) and several sub-Saharan African countries, including the Democratic Republic of Congo (Kamau et al. 2014; Mayengue et al. 2018), Uganda (Conrad et al. 2014), Equatorial Guinea (Li et al. 2016), Gabon (Kamau et al. 2014), Ghana (Matrevi et al. 2019), Kenya (Kamau et al. 2014), Togo (Dorkenoo et al. 2016), and Mali (Kamau et al. 2014; Dama et al. 2017). In vitro studies have confirmed that this mutation is not responsible for artemisinin resistance (Straimer et al. 2015; de Laurent et al. 2018; Ashley et al. 2014). However, because of its common occurrence in many countries, further characterization, as well as an assessment of its role in in vivo parasite clearance in sub-Saharan African countries is required. Moreover, the mutations P475H, Y482H, G665S, and A676S reported in our study have not been observed before. These results indicate that in Africa, parasites show a high polymorphism in pfk13 gene; however, the mutations observed are different from those reported in SEA. These differences seem to be affected by demographic and epidemiological parameters (Joy et al. 2003) rather than differential selective pressures that may be transient, and not necessarily due to artemisinin pressure (de Laurent et al. 2018). Although some candidate marker mutations have been reported in Africa, they were not involved in any case of treatment failure. Thus, the molecular mechanisms underlying artemisinin resistance remain unclear (Takala-Harrison et al. 2013). This phenomenon is likely to involve multiple genetic loci (Ariey et al. 2014; Miotto et al. 2015), and requires complete genotyping of circulating P. falciparum strains to detect new markers. Further studies, such as site-specific genome editing experiments using zinc finger nucleases (Mayengue et al. 2018), or the CRISPR-Cas9 system (Ghorbal et al. 2014), could shed light on the role of mutations discovered in sub-Saharan Africa, particularly in Côte d’Ivoire, in the resistance of P. falciparum to artemisinin derivatives.
Conclusion
This study showed that the adoption of ACTs as first-line treatment for uncomplicated malaria cases results in a significant change in the allele frequencies of molecular markers responsible for P. falciparum resistance against ASAQ and AL. For pfk13 gene, the mutations described in SEA and those associated with the resistance of P. falciparum to artemisinin derivatives were not observed in this study. An increase in the number of strains carrying the wild-type pfmdr1 N86 allele indicated lumefantrine tolerance. These results could throw the efficacy of artemether–lumefantrine into question, and thus demonstrate the importance of establishing a strong system for malarial molecular surveillance in Côte d’Ivoire.
References
Ako BA, Offianan AT, Johansson M et al (2012) Molecular analysis of markers associated with chloroquine and sulfadoxine/pyrimethamine resistance in Plasmodium falciparum Malaria parasites from southeastern Cote d’Ivoire by the time of artemisinin-based combination therapy adoption in 2005. Infect Drug Resist. https://doi.org/10.2147/IDR.S31409
Ariey F, Witkowski B, Amaratunga C et al (2014) A molecular marker of artemisinin-resistant Plasmodium falciparum Malaria. Nature 505:50–55. https://doi.org/10.1038/nature12876
Ashley EA, Dhorda M, Fairhurst RM et al (2014) Spread of artemisinin resistance in Plasmodium falciparum Malaria. N Engl J Med 371:411–423. https://doi.org/10.1056/NEJMoa1314981
Assi S-B, Henry M-C, Rogier C et al (2013) Inland Valley rice production systems and Malaria infection and disease in the forest region of western Côte d’Ivoire. Malar J 12:233. https://doi.org/10.1186/1475-2875-12-233
Azagoh-Kouadio R, Enoh S, Kassi K et al (2017) Paludisme De L’enfant: prise en charge Au Chu De Treichville. Rev Int Sci Médicales 19:26–30
Basco LK, Tahar R, Ako AB et al (2017) Molecular epidemiology of Malaria in Cameroon and Côte d’Ivoire. XXXI. Kelch 13 propeller sequences in Plasmodium falciparum isolates before and after implementation of artemisinin-based combination therapy. Am J Trop Med Hyg 97:222–224. https://doi.org/10.4269/ajtmh.16-0889
Bergmann C, van Loon W, Habarugira F et al (2021) Increase in Kelch 13 polymorphisms in Plasmodium falciparum, Southern Rwanda. Emerg Infect Dis 27:294–296. https://doi.org/10.3201/eid2701.203527
Bla BK, Yavo W, Trébissou J et al (2014) Polymorphisms of the pfatpase 6 and pfcrt gene and their relationship with the in vitro susceptibility to dihydroartemisinin and chloroquine of Plasmodium falciparum isolates from Abobo, Côte d’Ivoire. Ann Parasitol 60:259–266
Bwire GM, Ngasala B, Mikomangwa WP et al (2020) Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum Malaria in Southeast of Tanzania. Sci Rep 10:3500. https://doi.org/10.1038/s41598-020-60549-7
Conrad MD, Bigira V, Kapisi J et al (2014) Polymorphisms in K13 and Falcipain-2 Associated with Artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan Children. PLoS ONE 9:e105690. https://doi.org/10.1371/journal.pone.0105690
Dama S, Niangaly H, Ouattara A et al (2017) Reduced ex vivo susceptibility of Plasmodium falciparum after oral artemether–lumefantrine treatment in Mali. Malar J 16:59. https://doi.org/10.1186/s12936-017-1700-8
Dondorp AM, Fairhurst RM, Slutsker L et al (2011) The threat of artemisinin-resistant malaria. N Engl J Med 365:1073–1075. https://doi.org/10.1056/NEJMp1108322
Djaman J, Abouanou S, Basco L, Koné M (2004) Limits of the efficacy of chloroquine and sulfadoxine-pyrimethamine in Northern Abidjan (Cote d’Ivoire): combined in vivo and in vitro studies. Sante Montrouge Fr 14:205–209
Dokomajilar C, Nsobya SL, Greenhouse B et al (2006) Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where Malaria is highly endemic. Antimicrob Agents Chemother 50:1893–1895. https://doi.org/10.1128/AAC.50.5.1893-1895.2006
Dorkenoo AM, Yehadji D, Agbo YM et al (2016) Therapeutic efficacy trial of artemisinin-based combination therapy for the treatment of uncomplicated Malaria and investigation of mutations in k13 propeller domain in Togo, 2012–2013. Malar J. https://doi.org/10.1186/s12936-016-1381-8
Eyase FL, Akala HM, Ingasia L et al (2013) The role of Pfmdr1 and pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in Western-Kenya P. Falciparum samples during 2008–2011. PLoS ONE 8:e64299. https://doi.org/10.1371/journal.pone.0064299
Ghorbal M, Gorman M, Macpherson CR et al (2014) Genome editing in the human Malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. https://doi.org/10.1038/nbt.2925
Joy DA, Feng X, Mu J et al (2003) Early origin and recent expansion of lasmodium falciparum. Science 300:318–321. https://doi.org/10.1126/science.1081449
Kamau E, Campino S, Amenga-Etego L et al (2014) K13-Propeller polymorphisms in Plasmodium falciparum parasites from Sub-saharan Africa. J Infect Dis. https://doi.org/10.1093/infdis/jiu608
Konaté A, Barro-Kiki PCM, Angora KE et al (2018) Efficacy and tolerability of artesunate-amodiaquine versus artemether-lumefantrine in the treatment of uncomplicated Plasmodium falciparum Malaria at two sentinel sites across Côte d’Ivoire. Ann Parasitol 64:49–57
Konaté A, Gnagne PA, Bédia-Tanoh VA et al (2018) Low rates of Plasmodium falciparum Pfcrt K76T mutation in three sentinel sites of Malaria monitoring in Côte d’Ivoire. Acta Parasitol 63:795–801. https://doi.org/10.1515/ap-2018-0096
de Laurent ZR, Chebon LJ, Ingasia LA et al (2018) Polymorphisms in the K13 gene in Plasmodium falciparum from different Malaria transmission areas of Kenya. Am J Trop Med Hyg 98:1360–1366. https://doi.org/10.4269/ajtmh.17-0505
Li J, Chen J, Xie D et al (2016) Limited artemisinin resistance-associated polymorphisms in Plasmodium falciparum K13-propeller and PfATPase6 gene isolated from Bioko Island, Equatorial Guinea. Int J Parasitol Drugs Drug Resist 6:54–59. https://doi.org/10.1016/j.ijpddr.2015.11.002
Matrevi SA, Opoku-Agyeman P, Quashie NB et al (2019) Plasmodium Falciparum Kelch Propeller Polymorphisms in clinical isolates from Ghana from 2007 to 2016. Antimicrob Agents Chemother 63:e00802–e00819. https://doi.org/10.1128/AAC.00802-19
Mayengue PI, Niama RF, Kouhounina Batsimba D et al (2018) No polymorphisms in K13-propeller gene associated with artemisinin resistance in Plasmodium falciparum isolated from Brazzaville, Republic of Congo. BMC Infect Dis 18:538. https://doi.org/10.1186/s12879-018-3453-6
Menard S, Tchoufack JN, Maffo CN et al (2016) Insight into k13-propeller gene polymorphism and ex vivo DHA-response profiles from Cameroonian isolates. Malar J 15:572. https://doi.org/10.1186/s12936-016-1622-x
Ministère de la Santé et de l’Hygiène Publique (2007) Arrêté n°24/CAB/MSHP Du 12 janvier 2007 portant institution d’un schéma thérapeutique pour traitement du paludisme en Côte d’Ivoire. PNLP, Abidjan, Côte d’Ivoire
Ministère de la Santé et de la Lutte contre le SIDA (2015) Plan stratégique national de lutte contre le paludisme 2012–2015 révisé (période replanifiée): Approche stratifiée De mise à échelle des interventions de lutte contre le paludisme en Côte d’Ivoire et consolidation des acquis. PNLP, Abidjan, Côte d’Ivoire
Miotto O, Amato R, Ashley EA et al (2015) Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet 47:226–234. https://doi.org/10.1038/ng.3189
Noedl H, Se Y, Schaecher K et al (2008) Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359:2619–2620. https://doi.org/10.1056/NEJMc0805011
Okell LC, Reiter LM, Ebbe LS et al (2018) Emerging implications of policies on Malaria treatment: genetic changes in the Pfmdr-1 gene affecting susceptibility to artemether–lumefantrine and artesunate–amodiaquine in Africa. BMJ Glob Health 3:e000999. https://doi.org/10.1136/bmjgh-2018-000999
Ouattara L, Bla K, Assi S et al (2011) PFCRT and DHFR-TS sequences for monitoring drug resistance in Adzopé area of Côte d’Ivoire after the withdrawal of Chloroquine and Pyrimethamine. Trop J Pharm Res. https://doi.org/10.4314/tjpr.v9i6.63556
Plowe CV (2009) The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg 103:S11–S14. https://doi.org/10.1016/j.trstmh.2008.11.002
Raman J, Kagoro FM, Mabuza A et al (2019) Absence of kelch13 artemisinin resistance markers but strong selection for lumefantrine-tolerance molecular markers following 18 years of artemisinin-based combination therapy use in Mpumalanga Province, South Africa (2001–2018). Malar J 18:280. https://doi.org/10.1186/s12936-019-2911-y
Ringwald P, Sukwa T, Basco LK et al (2002) Monitoring of drug-resistant Malaria in Africa. Lancet 360:875–876. https://doi.org/10.1016/S0140-6736(02)09979-8
Sisowath C, Strömberg J, Mårtensson A et al (2005) In vivo selection of Plasmodium falciparum pfmdr1 86 N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis 191:1014–1017. https://doi.org/10.1086/427997
Sondo P, Derra K, Diallo Nakanabo S et al (2016) Artesunate-amodiaquine and artemether-lumefantrine therapies and selection of Pfcrt and Pfmdr1 alleles in Nanoro, Burkina Faso. PLoS ONE 11:e0151565. https://doi.org/10.1371/journal.pone.0151565
Straimer J, Gnädig NF, Witkowski B et al (2015) K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. https://doi.org/10.1126/science.1260867
Takala-Harrison S, Clark TG, Jacob CG et al (2013) Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci U S A 110:240–245. https://doi.org/10.1073/pnas.1211205110
Taylor SM, Parobek CM, DeConti DK et al (2015) Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-saharan Africa: a molecular epidemiologic study. J Infect Dis 211:680–688. https://doi.org/10.1093/infdis/jiu467
Toure OA, Assi SB, N’Guessan TL et al (2014) Open-label, randomized, non-inferiority clinical trial of artesunate-amodiaquine versus artemether-lumefantrine fixed-dose combinations in children and adults with uncomplicated falciparum Malaria in Côte d’Ivoire. Malar J 13:439. https://doi.org/10.1186/1475-2875-13-439
Trebissou JND, Yavo W, Tano KD et al (2014) In vitro susceptibility of Plasmodium falciparum to lumefantrine and analysis of polymophysims in pfmdr-1 gene isolates from Abidjan (Côte d’Ivoire). Int J Pharm Sci Res 5:1732–1738
Uwimana A, Legrand E, Stokes BH et al (2020) Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 26:1602–1608. https://doi.org/10.1038/s41591-020-1005-2
Uwimana A, Umulisa N, Venkatesan M et al (2021) Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect Dis 21:1120–1128. https://doi.org/10.1016/S1473-3099(21)00142-0
Venkatesan M, Gadalla NB, Stepniewska K et al (2014) Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. Falciparum Malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg 91:833–843. https://doi.org/10.4269/ajtmh.14-0031
Wang J, Huang Y, Zhao Y et al (2018) Introduction of F446I mutation in the K13 propeller gene leads to increased ring survival rates in Plasmodium falciparum isolates. Malar J 17:248. https://doi.org/10.1186/s12936-018-2396-0
WHO (2010) Basic malaria microscopy – Part I: Learner’s guide, 2nd edn. Switzerland, Geneva
WHO (2017) World Malaria report 2017. Geneva
Yavo W, Konaté A, Kassi FK et al (2015) Efficacy and safety of Artesunate-Amodiaquine versus Artemether-Lumefantrine in the treatment of uncomplicated Plasmodium falciparum malaria in sentinel sites across Côte d’Ivoire. Malar Res Treat 2015:1–8. https://doi.org/10.1155/2015/878132
Acknowledgements
We hereby thank the clinical study team and our administrative staff. We are also grateful to the patients who took part in this study.
Funding
Sample collection was a part of a study which received financial support from Global funds through the National Malaria Control Programme (NMCP) and National Institute of Public Health. The molecular analysis was supported by the Developing Excellence in Leadership and Genetics Training for Malaria Elimination (DELGEME) program in sub-Saharan Africa (Grant number: PD00217ML) through the DELTAS Africa Initiative (Grant number: 107740/Z/15/Z). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and is supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (Grant number: PD00217ML) and the United Kingdom government. Funders had no involvement in the study design, data collection, data analysis, data interpretation, manuscript drafting or the decision to submit it for publication.
Author information
Authors and Affiliations
Contributions
WY, EIHM supervised the study and corrected the manuscript. APG, AVBT and AKT supervised the samples collection. AKT and APG performed the molecular analysis. AKT and APG analysed the data and wrote this paper. All authors contributed to the drafting of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Konaté-Touré, A., Gnagne, A.P., Bedia-Tanoh, A.V. et al. Increase of Plasmodium falciparum parasites carrying lumefantrine-tolerance molecular markers and lack of South East Asian pfk13 artemisinin-resistance mutations in samples collected from 2013 to 2016 in Côte d’Ivoire. J Parasit Dis 48, 59–66 (2024). https://doi.org/10.1007/s12639-023-01640-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12639-023-01640-4