Abstract
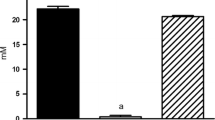
In the present study, we characterized the probiotic properties of two commercially available bacterial strains, Lactobacillus paragasseri UBLG-36 and Lacticaseibacillus paracasei UBLPC-87, and evaluated their ability to degrade oxalate in vitro and in a hyperoxaluria-induced nephrolithiasis rat model. UBLG-36 harboring two oxalate catabolizing genes, oxalyl coenzyme A decarboxylase (oxc) and formyl coenzyme A transferase (frc), was previously shown to degrade oxalate in vitro effectively. Here, we show that UBLPC-87, lacking both oxc and frc, could still degrade oxalate in vitro. Both these strains harbored several potential putative probiotic genes that may have conferred them the ability to survive in low pH and 0.3% bile, resist antibiotic stress, show antagonistic activity against pathogenic bacteria, and adhere to epithelial cell surfaces. We further evaluated if UBLG-36 and UBLPC-87 could degrade oxalate in vivo and prevent hyperoxaluria-induced nephrolithiasis in rats. We observed that rats treated with 4.5% sodium oxalate (NaOx) developed hyperoxaluria and renal stones. However, when pre-treated with UBLG-36 or UBLPC-87 before administering 4.5% NaOx, the rats were protected against several pathophysiological manifestations of hyperoxaluria. Compared to the hyperoxaluric rats, the probiotic pre-treated rats showed reduced urinary excretion of oxalate and urea (p < 0.05), decreased serum blood urea nitrogen and creatinine (p < 0.05), alleviated stone formation and renal histological damage, and an overall decrease in renal tissue oxalate and calcium content (p < 0.05). Taken together, both UBLG-36 and UBLPC-87 are effective oxalate catabolizing probiotics capable of preventing hyperoxaluria and alleviating renal damage associated with nephrolithiasis.








Similar content being viewed by others
Data Availability
All relevant data are within this article and its supplementary information files. The sequencing data presented in this article have been deposited to GenBank. L. paracasei UBLPC-87 whole-genome shotgun (WGS) project has the project accession JADDXY010000000 and consists of sequences JADDXY010000001-JADDXY010000021. The genome sequencing data of L. paragasseri UBLG-36 has been deposited in GenBank under the accession number JACOAE020000000 and consists of sequences JACOAE020000001-JACOAE020000005.
References
Demoulin N, Aydin S, Gillion V et al (2021) Pathophysiology and management of hyperoxaluria and oxalate nephropathy: a review. Am J Kidney Dis 79(5):717–727. https://doi.org/10.1053/J.AJKD.2021.07.018
Khan SR, Canales BK, Dominguez-Gutierrez PR (2021) Randall’s plaque and calcium oxalate stone formation: role for immunity and inflammation. Nat Rev Nephrol 17:417–433. https://doi.org/10.1038/S41581-020-00392-1
Worcester EM, Coe FL (2008) Nephrolithiasis. Prim Care Clin Off Pract 35:369–391. https://doi.org/10.1016/J.POP.2008.01.005
Gomathi S, Sasikumar P, Anbazhagan K et al (2014) Screening of indigenous oxalate degrading lactic acid bacteria from human faeces and South Indian fermented foods: assessment of probiotic potential. Sci World J 2014:1–11. https://doi.org/10.1155/2014/648059
Schwen ZR, Riley JM, Shilo Y, Averch TD (2013) Dietary management of idiopathic hyperoxaluria and the influence of patient characteristics and compliance. Urol 82:1220–1225. https://doi.org/10.1016/J.UROLOGY.2013.08.002
Siener R, Ebert D, Nicolay C, Hesse A (2003) Dietary risk factors for hyperoxaluria in calcium oxalate stone formers. Kidney Int 63:1037–1043. https://doi.org/10.1046/J.1523-1755.2003.00807.X
Nouvenne A, Meschi T, Guerra A et al (2009) Diet to reduce mild hyperoxaluria in patients with idiopathic calcium oxalate stone formation: a pilot study. Urol 73. https://doi.org/10.1016/J.UROLOGY.2008.11.006
Penniston KL, Nakada SY (2009) Effect of dietary changes on urinary oxalate excretion and calcium oxalate supersaturation in patients with hyperoxaluric stone formation. Urol 73:484–489. https://doi.org/10.1016/J.UROLOGY.2008.10.035
Pang R, Linnes MP, O’Connor HM et al (2012) Controlled metabolic diet reduces calcium oxalate supersaturation but not oxalate excretion after bariatric surgery. Urol 80:250–254. https://doi.org/10.1016/J.UROLOGY.2012.02.052
Hatch M (2017) Gut microbiota and oxalate homeostasis. Ann Transl Med 5:36. https://doi.org/10.21037/atm.2016.12.70
Liu M, Devlin JC, Hu J et al (2021) Microbial genetic and transcriptional contributions to oxalate degradation by the gut microbiota in health and disease. Elife 10:e63642. https://doi.org/10.7554/ELIFE.63642
Ticinesi A, Nouvenne A, Chiussi G et al (2020) Calcium oxalate nephrolithiasis and gut microbiota: not just a gut-kidney axis. a nutritional perspective. Nutrients 12(2):548. https://doi.org/10.3390/nu12020548
Suryavanshi MV, Bhute SS, Jadhav SD et al (2016) Hyperoxaluria leads to dysbiosis and drives selective enrichment of oxalate metabolizing bacterial species in recurrent kidney stone endures. Sci Rep 6:34712. https://doi.org/10.1038/srep34712
Ticinesi A, Milani C, Guerra A et al (2018) Understanding the gut-kidney axis in nephrolithiasis: an analysis of the gut microbiota composition and functionality of stone formers. Gut 67:2097–2106. https://doi.org/10.1136/GUTJNL-2017-315734
Miller AW, Choy D, Penniston KL, Lange D (2019) Inhibition of urinary stone disease by a multi-species bacterial network ensures healthy oxalate homeostasis. Kidney Int 96:180–188. https://doi.org/10.1016/j.kint.2019.02.012
Liu M, Koh H, Kurtz ZD et al (2017) Oxalobacter formigenes-associated host features and microbial community structures examined using the American Gut Project. Microbiome 5:108. https://doi.org/10.1186/s40168-017-0316-0
Mogna L, Pane M, Nicola S, Raiteri E (2014) Screening of different probiotic strains for their in vitro ability to metabolise oxalates. J Clin Gastroenterol 48:S91–S95. https://doi.org/10.1097/MCG.0000000000000228
Chamberlain CA, Hatch M, Garrett TJ (2019) Metabolomic profiling of oxalate-degrading probiotic Lactobacillus acidophilus and Lactobacillus gasseri. PLoS ONE 14:e0222393. https://doi.org/10.1371/journal.pone.0222393
Goldstein EJC, Tyrrell KL, Citron DM (2015) Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis 60:S98–S107. https://doi.org/10.1093/CID/CIV072
Sanders ME, Merenstein DJ, Reid G et al (2019) Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol 1610(16):605–616. https://doi.org/10.1038/s41575-019-0173-3
Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A (2019) Mechanisms of action of probiotics. Adv Nutr 10:S49–S66. https://doi.org/10.1093/ADVANCES/NMY063
Hempel S, Newberry SJ, Maher AR et al (2012) Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA 307:1959–1969. https://doi.org/10.1001/JAMA.2012.3507
Moayyedi P, Ford AC, Talley NJ et al (2010) The efficacy of probiotics in the treatment of irritable bowel syndrome: a systematic review. Gut 59:325–332. https://doi.org/10.1136/GUT.2008.167270
Goldenberg JZ, Yap C, Lytvyn L et al (2017) Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev 12(12):CD006095. https://doi.org/10.1002/14651858.CD006095.PUB4
Saez-Lara MJ, Gomez-Llorente C, Plaza-Diaz J, Gil A (2015) The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: a systematic review of randomized human clinical trials. Biomed Res Int 2015:505878. https://doi.org/10.1155/2015/505878
Koutnikova H, Genser B, Monteiro-Sepulveda M et al (2019) Impact of bacterial probiotics on obesity, diabetes and non-alcoholic fatty liver disease related variables: a systematic review and meta-analysis of randomised controlled trials. BMJ Open 9:e017995. https://doi.org/10.1136/BMJOPEN-2017-017995
Kocsis T, Molnár B, Németh D et al (2020) (2020) Probiotics have beneficial metabolic effects in patients with type 2 diabetes mellitus: a meta-analysis of randomized clinical trials. Sci Reports 101(10):1–14. https://doi.org/10.1038/s41598-020-68440-1
da Silva TF, Casarotti SN, de Oliveira GLV, Penna ALB (2020) The impact of probiotics, prebiotics, and synbiotics on the biochemical, clinical, and immunological markers, as well as on the gut microbiota of obese hosts. Crit Rev Food Sci Nutr 61(2):337–355. https://doi.org/10.1080/1040839820201733483
Wigner P, Bijak M, Saluk-Bijak J (2022) Probiotics in the prevention of the calcium oxalate urolithiasis. Cells (2022) Vol 11. Page 284(11):284. https://doi.org/10.3390/CELLS11020284
Turroni S, Vitali B, Bendazzoli C et al (2007) Oxalate consumption by lactobacilli: evaluation of oxalyl-CoA decarboxylase and formyl-CoA transferase activity in Lactobacillus acidophilus. J Appl Microbiol 103:1600–1609. https://doi.org/10.1111/j.1365-2672.2007.03388.x
Murphy C, Murphy S, O’Brien F et al (2009) Metabolic activity of probiotics-oxalate degradation. Vet Microbiol 136:100–107. https://doi.org/10.1016/j.vetmic.2008.10.005
Hatch M (2017) Gut microbiota and oxalate homeostasis. Ann Transl Med 5:36–36. https://doi.org/10.21037/atm.2016.12.70
Mehra Y, Viswanathan P (2021) High-quality whole-genome sequence analysis of Lactobacillus paragasseri UBLG-36 reveals oxalate-degrading potential of the strain. PLoS One 16(11):e0260116. https://doi.org/10.1371/JOURNAL.PONE.0260116
Campieri C, Campieri M, Bertuzzi V et al (2001) Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int 60:1097–1105. https://doi.org/10.1046/j.1523-1755.2001.0600031097.x
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol 13:e1005595. https://doi.org/10.1371/journal.pcbi.1005595
Bosi E, Donati B, Galardini M et al (2015) MeDuSa: a multi-draft based scaffolder. Bioinformatics 31:2443–2451. https://doi.org/10.1093/bioinformatics/btv171
Tatusova T, Dicuccio M, Badretdin A et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1093/nar/gkw569
Cosentino S, Voldby Larsen M, Møller Aarestrup F, Lund O (2013) PathogenFinder - distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 8:e77302. https://doi.org/10.1371/JOURNAL.PONE.0077302
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. https://doi.org/10.1093/bioinformatics/btv681
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:1–10. https://doi.org/10.1038/s41467-019-10210-3
Fernández MF, Boris S, Barbés C (2003) Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J Appl Microbiol 94:449–455. https://doi.org/10.1046/j.1365-2672.2003.01850.x
Rychen G, Aquilina G, Azimonti G et al (2018) Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J 16(30):5206. https://doi.org/10.2903/J.EFSA.2018.5206
Humphries R, Bobenchik AM, Hindler JA, Schuetz AN (2021) Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st Edition. J Clin Microbiol 59:e0021321. https://doi.org/10.1128/JCM.00213-21
Hu CH, Ren LQ, Zhou Y, Ye BC (2019) Characterization of antimicrobial activity of three Lactobacillus plantarum strains isolated from Chinese traditional dairy food. Food Sci Nutr 7:1997–2005. https://doi.org/10.1002/fsn3.1025
Prabhurajeshwar C, Chandrakanth K (2019) Evaluation of antimicrobial properties and their substances against pathogenic bacteria in-vitro by probiotic lactobacilli strains isolated from commercial yoghurt. Clin Nutr Exp 23:97–115. https://doi.org/10.1016/j.yclnex.2018.10.001
Mishra AK, Ghosh AR (2018) Characterization of functional, safety, and probiotic properties of Enterococcus faecalis AG5 isolated from Wistar rat, demonstrating adherence to HCT 116 cells and gastrointestinal survivability. Probiotics Antimicrob Proteins 10:435–445. https://doi.org/10.1007/S12602-018-9387-X
Ducret A, Quardokus EM, Brun YV (2016) MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. https://doi.org/10.1038/NMICROBIOL.2016.77
Schindelin J, Arganda-Carreras I, Frise E et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 97(9):676–682. https://doi.org/10.1038/nmeth.2019
Hodgkinson A, Williams A (1972) An improved colorimetric procedure for urine oxalate. Clin Chim Acta 36:127–132. https://doi.org/10.1016/0009-8981(72)90167-2
Gomathi S, Sasikumar P, Anbazhagan K et al (2015) Oral administration of indigenous oxalate degrading lactic acid bacteria and quercetin prevents calcium oxalate stone formation in rats fed with oxalate rich diet. J Funct Foods 17:43–54. https://doi.org/10.1016/j.jff.2015.05.011
Kapse NG, Engineer AS, Gowdaman V et al (2019) Functional annotation of the genome unravels probiotic potential of Bacillus coagulans HS243. Genomics 111:921–929. https://doi.org/10.1016/J.YGENO.2018.05.022
Zhang W, Wang J, Zhang D et al (2019) Complete genome sequencing and comparative genome characterization of Lactobacillus johnsonii ZLJ010, a potential probiotic with health-promoting properties. Front Genet 10:812. https://doi.org/10.3389/fgene.2019.00812
Nguyen TTN, Seo E, Choi J et al (2017) Phosphatidylinositol 4-phosphate 5-kinase α contributes to Toll-like receptor 2-mediated immune responses in microglial cells stimulated with lipoteichoic acid. Cell Signal 38:159–170. https://doi.org/10.1016/j.cellsig.2017.07.009
Yang L, Li W, Ujiroghene OJ et al (2020) Occurrence and diversity of CRISPR loci in Lactobacillus casei group. Front Microbiol 11:642. https://doi.org/10.3389/FMICB.2020.00624
Shehata MG, El Sohaimy SA, El-Sahn MA, Youssef MM (2016) Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann Agric Sci 61:65–75. https://doi.org/10.1016/J.AOAS.2016.03.001
Silva DR, Sardi JD, de Souza Pitangui N, Pitangui N et al (2020) Probiotics as an alternative antimicrobial therapy: current reality and future directions. J Funct Foods 73. https://doi.org/10.1016/J.JFF.2020.104080
Raheem A, Liang L, Zhang G, Cui S (2021) Modulatory effects of probiotics during pathogenic infections with emphasis on immune regulation. Front Immunol 12:571. https://doi.org/10.3389/FIMMU.2021.616713/BIBTEX
Meroni G, Panelli S, Zuccotti G et al (2021) Probiotics as therapeutic tools against pathogenic biofilms: have we found the perfect weapon? Microbiol Res 12:916–937. https://doi.org/10.3390/MICROBIOLRES12040068
Ibrahim SA, Ayivi RD, Zimmerman T et al (2021) Lactic acid bacteria as antimicrobial agents: food safety and microbial food spoilage prevention. Foods 10(12):3131. https://doi.org/10.3390/FOODS10123131
Lea T (2015) Caco-2 cell line. Impact Food Bioact Heal Vitr Ex Vivo Model 103–111. https://doi.org/10.1007/978-3-319-16104-4_10
Ley RE, Hamady M, Lozupone C et al (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651. https://doi.org/10.1126/SCIENCE.1155725/SUPPL_FILE/LEY.SOM.PDF
Gnanandarajah JS, Johnson TJ, Kim HB et al (2012) Comparative faecal microbiota of dogs with and without calcium oxalate stones. J Appl Microbiol 113:745–756. https://doi.org/10.1111/J.1365-2672.2012.05390.X
Kwak C, Jeong BC, Ku JH et al (2006) Prevention of nephrolithiasis by Lactobacillus in stone-forming rats: a preliminary study. Urol Res 34:265–270. https://doi.org/10.1007/S00240-006-0054-4
Kaufman DW, Kelly JP, Curhan GC et al (2008) Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol 19:1197–1203. https://doi.org/10.1681/ASN.2007101058
Miller AW, Oakeson KF, Dale C, Dearing MD (2016) Effect of dietary oxalate on the gut microbiota of the mammalian herbivore Neotoma albigula. Appl Environ Microbiol 82:2669–2675. https://doi.org/10.1128/AEM.00216-16
Shirley EK, Schmidt-Nielsen K (1967) Oxalate metabolism in the pack rat, sand rat, hamster, and white rat. J Nutr 91:496–502. https://doi.org/10.1093/JN/91.4.496
Lieske JC, Tremaine WJ, De Simone C et al (2010) Diet, but not oral probiotics, effectively reduces urinary oxalate excretion and calcium oxalate supersaturation. Kidney Int 78:1178–1185. https://doi.org/10.1038/ki.2010.310
Miller AW, Dale C, Dearing MD (2017) Microbiota diversification and crash induced by dietary oxalate in the mammalian herbivore Neotoma albigula. mSphere 2:e00428-17. https://doi.org/10.1128/MSPHERE.00428-17/SUPPL_FILE/SPH005172383ST7.PDF
Yamaguchi S, Wiessner JH, Hasegawa AT et al (2005) Study of a rat model for calcium oxalate crystal formation without severe renal damage in selected conditions. Int J Urol 12:290–298. https://doi.org/10.1111/J.1442-2042.2005.01038.X
Bushinsky DA, Bashir MA, Riordon DR et al (1999) Increased dietary oxalate does not increase urinary calcium oxalate saturation in hypercalciuric rats. Kidney Int 55:602–612. https://doi.org/10.1046/j.1523-1755.1999.00281.x
Stewart CS, Duncan SH, Cave DR (2004) Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol Lett 230:1–7. https://doi.org/10.1016/S0378-1097(03)00864-4
Huang Y, Zhang YH, Chi ZP et al (2020) The handling of oxalate in the body and the origin of oxalate in calcium oxalate stones. Urol Int 104:167–176. https://doi.org/10.1159/000504417
Giardina S, Scilironi C, Michelotti A et al (2014) In vitro anti-inflammatory activity of selected oxalate-degrading probiotic bacteria: potential applications in the prevention and treatment of hyperoxaluria. J Food Sci 79:384–390. https://doi.org/10.1111/1750-3841.12344
Sasikumar P, Gomathi S, Anbazhagan K et al (2014) Recombinant Lactobacillus plantarum expressing and secreting heterologous oxalate decarboxylase prevents renal calcium oxalate stone deposition in experimental rats. J Biomed Sci 21:86. https://doi.org/10.1186/S12929-014-0086-Y
Klimesova K, Whittamore JM, Hatch M (2015) Bifidobacterium animalis subsp. lactis decreases urinary oxalate excretion in a mouse model of primary hyperoxaluria. Urolithiasis 43:107–117. https://doi.org/10.1007/s00240-014-0728-2
Abratt VR, Reid SJ (2010) Oxalate-degrading bacteria of the human gut as probiotics in the management of kidney stone disease. Adv Appl Microbiol 72:63–87. https://doi.org/10.1016/S0065-2164(10)72003-7
Papadimitriou K, Zoumpopoulou G, Foligné B, et al (2015) Discovering probiotic microorganisms: in vitro, in vivo, genetic and omics approaches. Front Microbiol 6:1–28. https://doi.org/10.3389/fmicb.2015.00058
Papadimitriou K, Alegría Á, Bron PA et al (2016) Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev 80:837. https://doi.org/10.1128/MMBR.00076-15
Maresca D, Zotta T, Mauriello G (2018) Adaptation to aerobic environment of Lactobacillus johnsonii/gasseri strains. Front Microbiol 9:157. https://doi.org/10.3389/FMICB.2018.00157/FULL
Fuochi V, Petronio GP, Lissandrello E, Furneri PM (2015) Evaluation of resistance to low pH and bile salts of human Lactobacillus spp. isolates. Int J Immunopathol Pharmacol 28:426–433. https://doi.org/10.1177/0394632015590948
Mallappa RH, Singh DK, Rokana N et al (2019) Screening and selection of probiotic Lactobacillus strains of Indian gut origin based on assessment of desired probiotic attributes combined with principal component and heatmap analysis. LWT 105:272–281. https://doi.org/10.1016/J.LWT.2019.02.002
Anisimova EA (2019) Yarullina DR (2019) Antibiotic resistance of Lactobacillus strains. Curr Microbiol 7612(76):1407–1416. https://doi.org/10.1007/S00284-019-01769-7
Bacha K, Mehari T, Ashenafi M (2010) Antimicrobial susceptibility patterns of lab isolated from wakalim, a traditional Ethiopian fermented sausage. J Food Saf 30:213–223. https://doi.org/10.1111/J.1745-4565.2009.00201.X
Li T, Teng D, Mao R et al (2020) A critical review of antibiotic resistance in probiotic bacteria. Food Res Int 136:109571. https://doi.org/10.1016/J.FOODRES.2020.109571
Gueimonde M, Sánchez B, de los Reyes-Gavilán CG, Margolles A, (2013) Antibiotic resistance in probiotic bacteria. Front Microbiol 4:202. https://doi.org/10.3389/FMICB.2013.00202/BIBTEX
Sharma C, Gulati S, Thakur N et al (2017) Antibiotic sensitivity pattern of indigenous lactobacilli isolated from curd and human milk samples. 3 Biotech 7:53. https://doi.org/10.1007/S13205-017-0682-0
Kumar A, Kumar D (2015) Characterization of Lactobacillus isolated from dairy samples for probiotic properties. Anaerobe 33:117–123. https://doi.org/10.1016/J.ANAEROBE.2015.03.004
Imperial ICVJ, Ibana JA (2016) Addressing the antibiotic resistance problem with probiotics: reducing the risk of its double-edged sword effect. Front Microbiol 7:1983. https://doi.org/10.3389/FMICB.2016.01983
Elkins CA, Mullis LB (2004) Bile-mediated aminoglycoside sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl Environ Microbiol 70:7200–7209. https://doi.org/10.1128/AEM.70.12.7200-7209.2004
Liasi SA, Azmi H et al (2009) Antimicrobial activity and antibiotic sensitivity of three isolates of lactic acid bacteria from fermented fish product, Budu. Malays J Microbiol 5:33–37
Mathur S, Singh R (2005) Antibiotic resistance in food lactic acid bacteria–a review. Int J Food Microbiol 105:281–295. https://doi.org/10.1016/J.IJFOODMICRO.2005.03.008
Fao J, Working WHO, Report G et al (2002) Guidelines for the evaluation of probiotics in food. 1–11
Bermudez-Brito M, Plaza-Díaz J, Muñoz-Quezada S et al (2012) Probiotic mechanisms of action. Ann Nutr Metab 61:160–174. https://doi.org/10.1159/000342079
Gaspar C, Donders GG, Palmeira-de-Oliveira R et al (2018) Bacteriocin production of the probiotic Lactobacillus acidophilus KS400. AMB Express 8:153. https://doi.org/10.1186/S13568-018-0679-Z
Makras L, Triantafyllou V, Fayol-Messaoudi D et al (2006) Kinetic analysis of the antibacterial activity of probiotic lactobacilli towards Salmonella enterica serovar Typhimurium reveals a role for lactic acid and other inhibitory compounds. Res Microbiol 157:241–247. https://doi.org/10.1016/J.RESMIC.2005.09.002
De Keersmaecker SCJ, Verhoeven TLA, Desair J et al (2006) Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol Lett 259:89–96. https://doi.org/10.1111/J.1574-6968.2006.00250.X
Collado MC, Meriluoto J, Salminen S (2007) Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett Appl Microbiol 45:454–460. https://doi.org/10.1111/J.1472-765X.2007.02212.X
Chen CC, Lai CC, Huang HL et al (2019) Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front Microbiol 10:789. https://doi.org/10.3389/FMICB.2019.00789/BIBTEX
Duary RK, Rajput YS, Batish VK, Grover S (2011) Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian J Med Res 134:664–671. https://doi.org/10.4103/0971-5916.90992
Archer AC, Kurrey NK, Halami PM (2018) In vitro adhesion and anti-inflammatory properties of native Lactobacillus fermentum and Lactobacillus delbrueckii spp. J Appl Microbiol 125:243–256. https://doi.org/10.1111/JAM.13757
Coe FL, Worcester EM, Evan AP (2016) Idiopathic hypercalciuria and formation of calcium renal stones. Nat Rev Nephrol 12:519. https://doi.org/10.1038/NRNEPH.2016.101
Moe OW (2006) Kidney stones: pathophysiology and medical management. Lancet 367:333–344. https://doi.org/10.1016/S0140-6736(06)68071-9
Paul E, Albert A, Ponnusamy S et al (2018) Designer probiotic Lactobacillus plantarum expressing oxalate decarboxylase developed using group II intron degrades intestinal oxalate in hyperoxaluric rats. Microbiol Res 215:65–75. https://doi.org/10.1016/j.micres.2018.06.009
McMartin K (2009) Are calcium oxalate crystals involved in the mechanism of acute renal failure in ethylene glycol poisoning ethylene glycol renal toxicity mechanism. Clin Toxicol 47:859–869. https://doi.org/10.3109/15563650903344793
Johansson G, Backman U, Danielson BG et al (1982) Effects of magnesium hydroxide in renal stone disease. J Am Coll Nutr 1:179–185. https://doi.org/10.1080/07315724.1982.10718985
Taylor EN, Hoofnagle AN, Curhan GC (2015) Calcium and phosphorus regulatory hormones and risk of incident symptomatic kidney stones. Clin J Am Soc Nephrol 10:667–675. https://doi.org/10.2215/CJN.07060714
Baumann JM (2014) From crystalluria to kidney stones, some physicochemical aspects of calcium nephrolithiasis. World J Nephrol 3:256. https://doi.org/10.5527/wjn.v3.i4.256
Lieske JC, Goldfarb DS, De Simone C, Regnier C (2005) Use of a probiotic to decrease enteric hyperoxaluria. Kidney Int 68:1244–1249. https://doi.org/10.1111/j.1523-1755.2005.00520.x
Acknowledgements
The authors thank the Vellore Institute of Technology (VIT) for providing all necessary research amenities. The authors are also grateful to Unique Biotech Pvt. Ltd. for providing the bacteria. We highly appreciate Illume Gene India LLP for their assistance in sequencing the bacterial strains.
Funding
This study was supported by the Indian Council of Medical Research (ICMR), New Delhi (Grant No. 5/9/1094/2013-Nut).
Author information
Authors and Affiliations
Contributions
Conceptualization: Yogita Mehra and Pragasam Viswanathan. Data curation: Pragasam Viswanathan. Methodology: Yogita Mehra. Formal analysis: Yogita Mehra. Histopathology interpretation: Rajesh NG. Investigation: Yogita Mehra. Writing—original draft: Yogita Mehra. Writing—review and editing: Yogita Mehra and Pragasam Viswanathan. Supervision: Pragasam Viswanathan. Resources: Pragasam Viswanathan. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical Approval
The study was approved by the Institutional Animal Ethics Committee (Registration No 1333/c/10/CPCSEA; Approval Number-VIT/IAEC/15/Sep2/25). Experiments were carried out according to the guidelines set by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mehra, Y., Rajesh, N.G. & Viswanathan, P. Analysis and Characterization of Lactobacillus paragasseri and Lacticaseibacillus paracasei: Two Probiotic Bacteria that Can Degrade Intestinal Oxalate in Hyperoxaluric Rats. Probiotics & Antimicro. Prot. 14, 854–872 (2022). https://doi.org/10.1007/s12602-022-09958-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-022-09958-w