Abstract
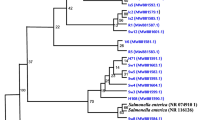
The bacterial isolates from Centroscyllium fabricii (deep sea shark) gut were screened for antagonistic activity by cross-streak method and disc diffusion assay. This study focuses on strain BTSS-3, which showed antimicrobial activity against pathogenic bacteria including Salmonella Typhimurium, Proteus vulgaris, Clostridium perfringens, Staphylococcus aureus, Bacillus cereus, Bacillus circulans, Bacillus macerans and Bacillus pumilus. BTSS3 was subjected to phenotypic characterization using biochemical tests, SEM imaging, exoenzyme profiling and antibiotic susceptibility tests. Comparative 16S rDNA gene sequence analysis indicated that this strain belonged to the genus Bacillus, with high (98 %) similarity to 16S rDNA sequences of Bacillus amyloliquefaciens. The chemical nature of the antibacterial substance was identified by treatment with proteolytic enzymes. The antibacterial activity was reduced by the action of these enzymes pointing out its peptide nature. It was observed from the growth and production kinetics that the bacteriocin was produced in the eighth hour of incubation, i.e., during the mid-log growth phase of the bacteria.



Similar content being viewed by others
References
De Vuyst L, Vandamme EJ (1994) Bacteriocins of lactic acid bacteria: microbiology, genetics and applications. London, Blackie Academic & Professional, Genetics and Applications
De Vuyst L, Leroy F (2007) Bacteriocins from lactic acid bacteria: production, purification, and food applications. J Mol Microbiol Biotechnol 13:194–199
Lisboa MP, Bonatto D, Bizani D, Henriques JAP, Brandelli A (2006) Characterization of a bacteriocin-like substance produced by Bacillus amyloliquefaciens isolated from the Brazilian Atlantic Forest. Int Microbiol 9:111–118
O’Sullivan L, Ross RP, Hill C (2002) Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. Biochimie 84:593–604
Riley MA, Gordon DM (1992) A survey of col plasmids in natural isolates of Escherichia coli and an investigation into the stability of col-plasmid lineages. J Microbiol 138:1345–1352
James R, Lazdunski C, Pattus F (eds) (1991) Bacteriocins, microcins and lantibiotics, vol 65. Springer, New York
Olafsen JA (2001) Interactions between fish larvae and bacteria in marine aquaculture. Aquaculture 200:223–247
Westerdahl A, Olsson JC, Kjelleberg S, Conway P (1991) Isolation and Characterization of Turbot (Scophtalmus maximus)-associated bacteria with inhibitory effects against Vibrio anguillarum. Appl Environ Microbiol 57:2223–2228
Ringø E, Bendiksen HR, Wesmajervi MS, Olsen RE, Jansen PA, Mikkelsen H (2000) Lactic acid bacteria associated with the digestive tract of Atlantic salmon (Salmo salar L.). J Appl Microbiol 89:317–322
Makridis P, Martins S, Tsalavouta M, Dionisio LC, Kotoulas G, Magoulas A, Dinis MT (2005) Antimicrobial activity in bacteria isolated from Senegalese sole, Solea senegalensis, fed with natural prey. Aquacult Res 36:1619–1627
Sugita H, Shibuya K, Hanada H, Deguchi Y (1997) Antibacterial abilities of intestinal microflora of the river fish. Fish Sci 63:378–383
Bergh Ø (1995) Bacteria associated with early-life stages of halibut, Hippoglossus hippoglossus, inhibit growth of a pathogenic Vibrio sp. J Fish Dis 18:31–40
Gardner JF (1950) Some antibiotics formed by Bacterium coli. Br J Exp Pathol 31:102–111
Bauer AW, Kirby WMM, Sherris JC, Truck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193:265–275
Heyns W, De Moor P (1971) The binding of 17β-hydroxy-5α-androstan-3-one to the steroid-binding β-globulin in human plasma, as studied by means of ammonium sulphate precipitation. Steroids 18:709–730
Mayr-Harting A, Hedges AJ, Berkeley CW (1972) Methods for studying bacteriocins. Methods Microbiol 7:315–412
Enan G, El-Essway AA, Uyttendaele M, Debevere J (1996) Antibacterial activity of Lactobacillus plantarum UG1 isolated from dry sausage: characterization, production and bactericidal action of plantaricin UG1. Int J Food Microbiol 30:189–215
Skerman VBD (1967) A guide to the identification of the genera of bacteria, 2nd edn. Williams & Wilkins, Baltimore
Dong XZ, Cai MY (2001) Determinative manual for routine bacteriology. Scientific Press (English translation), Beijing
Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K (1987) Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley Interscience, New York
Shivaji S, Bhanu NV, Aggarwal RK (2000) Identification of Yersinia pestis as the causative organism of plague in India as determined by 16SrDNA sequencing and RAPD-based genomic fingerprinting. FEMS Microbiol Lett 189:247–252
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Parkes RJ, Cragg BA, Bale SJ, Getlifff JM, Goodman K, Rochelle PA, Fry JC, Weightman AJ, Harvey SM (1994) Deep bacterial biosphere in Pacific Ocean sediments. Nature 371:410–413. doi:10.1038/371410a0
Sugita H, Ushioka S, Kihara D, Deguchi Y (1985) Changes in the bacterial composition of water in a carp rearing tank. Aquaculture 44:243–247
Smith P, Davey S (1993) Evidence for the competitive exclusion of Aeromonas salmonicida from fish with stress-inducible furunculosis by a fluorescent pseudomonad. J Fish Dis 16:521–524
Austin B, Stuckey LF, Robertson PAW, Effendi I, Griffith DRW (1995) A probiotic strain of Vibrio alginolyticus effective in reducing diseases caused by Aeromonas salmonicida, Vibrio anguillarum and Vibrio ordalii. J Fish Dis 18:93–96
Cladera-Olivera F, Caron GR, Brandelli A (2004) Bacteriocin-like substance production by Bacillus licheniformis strain P40. Lett Appl Microbiol 38:251–256
Hoyt PR, Sizemore RK (1982) Competitive dominance by a bacteriocin-producing Vibrio harveyi strain. Appl Environ Microbiol 44:653–658
Nicholson WL (2002) Roles of Bacillus endospores in the environment. Cell Mol Life Sci 59:410–416
Sutyak KE, Wirawan RE, Aroutcheva AA, Chikindas ML (2008) Isolation of the Bacillus subtilis antimicrobial peptide subtilosin from the dairy product-derived Bacillus amyloliquefaciens. J Appl Microbiol 104:1067–1074
Lawton EM, Paul D, Cotter CH, Ross PR (2006) Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett 267:64–71
Gray EJ, Lee KD, Souleimanov AM, Di Falco MR, Zhou X, Ly A, Charles TC, Driscoll BT et al (2006) A novel bacteriocin, thuricin 17, produced by plant growth promoting rhizobacteria strain Bacillus thuringiensis NEB17: isolation and identification. J Appl Microbiol 100:545–554
Martirani L, Varcamonti M, Naclerio G, Felice MD (2002) Purification and partial characterization of bacillocin 490, a novel bacteriocin produced by a thermophilic strain of Bacillus licheniformis. Microb Cell Fact. doi:10.1186/1475-2859-1-1
Oscariz JC, Lasa I, Pissabarro AG (1999) Detection and characterization of cerien 7, a new bacteriocin produced by Bacillus cereus with a broad spectrum of activity. FEMS Microbiol Lett 178:337–341
Banerjee S, Hansen JN (1988) Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J Biol Chem 263:9508–9514
Reiss R, Ihssen J, Thöny-Meyer L (2011) Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum. BMC Biotechnol 11:9. doi:10.1186/1472-6750-11-9
Mukherjee S, Das P, Sivapathasekaran C, Sen R (2009) Antimicrobial biosurfactants from marine Bacillus circulans: extracellular synthesis and purification. Lett Appl Microbiol 48:281–288
Parente E, Ricciardi A (1994) Influence of pH on the production of enterocin 1146 during batch fermentation. Lett Appl Microbiol 19:12–15
De Vuyst L, Callewaert R, Crabbe K (1996) Primary metabolite kinetics of bacteriocin biosynthesis by Lactobacillus amylovorus and evidence for stimulation of bacteriocin production under unfavourable growth conditions. Microbiology 142:817–827
Smitha S, Bhat SG (2013) Thermostable Bacteriocin BL8 from Bacillus licheniformis isolated from marine sediment. J Appl Microbiol 114:688–694
Naclerio G, Ricca E, Sacco M, De Felice M (1993) Antimicrobial activity of a newly identified bacteriocin of Bacillus cereus. Appl Environ Microbiol 59:4313–4316
Acknowledgments
This study was supported by project Grants from Centre for Marine Living Resources and Ecology- Ministry of Earth Sciences, Government of India (MOES/10-MLR/2/2007 and MOES/10-MLR-TD/03/2013) given to Dr. Sarita G. Bhat, Department of Biotechnology, Cochin University of Science and Technology, Kochi, India.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
This article does not contain any studies with animals or human participants performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bindiya, E.S., Tina, K.J., Raghul, S.S. et al. Characterization of Deep Sea Fish Gut Bacteria with Antagonistic Potential, from Centroscyllium fabricii (Deep Sea Shark). Probiotics & Antimicro. Prot. 7, 157–163 (2015). https://doi.org/10.1007/s12602-015-9190-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-015-9190-x