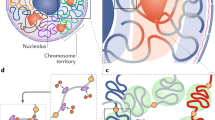
Abstract
Chromatin exists as a non-linear, “three-dimensional” structure in the nuclear space. The dynamic alteration of the chromatin structure leads to transcriptional changes during the formation of the neuronal network. Several studies providing evidence for the link between the dysregulation of spatial chromatin architecture and developmental disorders have accumulated. Therefore, we studied and reviewed the regulation and dysregulation of 3D genome organization in the central nervous system, with a special focus on the cohesin complex that is crucial for the formation of the chromatin loop structure. This review summarizes the function and mechanisms of spatial chromatin architecture during the development of the central nervous system. We discuss the link between the disturbances in the 3D chromatin structure and the diseases of the central nervous system. Finally, we discuss how the knowledge of 3D genome organization may lead to further advances in diagnosis and therapy for the diseases of the central nervous system.





Similar content being viewed by others
References
Beagan JA, Pastuzyn ED, Fernandez LR et al (2020) Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression. Nat Neurosci 23:707–717
Bickmore WA, Van Steensel B (2013) Genome architecture: domain organization of interphase chromosomes. Cell 152:1270–1284
Billia F, Baskys A, Carlen PL, De Boni U (1992) Rearrangement of centromeric satellite DNA in hippocampal neurons exhibiting long-term potentiation. Brain Res Mol Brain Res 14:101–108
Bonev B, Cavalli G (2016) Organization and function of the 3D genome. Nat Rev Genet 17:772
Bonev B, Mendelson Cohen N, Szabo Q et al (2017) Multiscale 3D genome rewiring during mouse neural development. Cell 171(557–572):e24
Castronovo P, Gervasini C, Cereda A et al (2009) Premature chromatid separation is not a useful diagnostic marker for Cornelia de Lange syndrome. Chromosome Res 17:763–771
Cavalli G, Misteli T (2013) Functional implications of genome topology. Nat Struct Mol Biol 20:290–299
Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM (2019) DNA loop extrusion by human cohesin. Science 366:1338–1345
De Lange C (1933) Sur un type nouveau de dégénératio. Arch Méd Enfants 36:713–719
De Laat W, Duboule D (2013) Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502:499–506
Deardorff MA, Kaur M, Yaeger D et al (2007) Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet 80:485–494
Deardorff MA, Bando M, Nakato R et al (2012a) HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489:313–317
Deardorff MA, Wilde JJ, Albrecht M et al (2012b) RAD21 mutations cause a human cohesinopathy. Am J Hum Genet 90:1014–1027
Dekker J, Heard E (2015) Structural and functional diversity of Topologically Associating Domains. FEBS Lett 589:2877–2884
Dixon JR, Selvaraj S, Yue F et al (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380
Dixon JR, Gorkin DU, Ren B (2016) Chromatin domains: the unit of chromosome organization. Mol Cell 62:668–680
Feodorova Y, Falk M, Mirny LA, Solovei I (2020) Viewing nuclear architecture through the eyes of nocturnal mammals. Trends Cell Biol 30:276–289
Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA (2016) Formation of chromosomal domains by loop extrusion. Cell Rep 15:2038–2049
Fujita Y, Yamashita T (2018) Spatial organization of genome architecture in neuronal development and disease. Neurochem Int 119:49–56
Fujita Y, Masuda K, Bando M et al (2017) Decreased cohesin in the brain leads to defective synapse development and anxiety-related behavior. J Exp Med 214:1431–1452
Ganji M, Shaltiel IA, Bisht S et al (2018) Real-time imaging of DNA loop extrusion by condensin. Science 360:102–105
Gibcus JH, Dekker J (2013) The hierarchy of the 3D genome. Mol Cell 49:773–782
Gorkin DU, Leung D, Ren B (2014) The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 14:762–775
Guerreiro I, Kind J (2019) Spatial chromatin organization and gene regulation at the nuclear lamina. Curr Opin Genet Dev 55:19–25
Guo Y, Xu Q, Canzio D et al (2015) CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162:900–910
Hadjur S, Williams LM, Ryan NK et al (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460:410–413
Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X (2017) CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 6:e25776
Hao N, Shearwin KE, Dodd IB (2017) Programmable DNA looping using engineered bivalent dCas9 complexes. Nat Commun 8:1628
Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T (2012) CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep 2:345–357
Ireland M, Donnai D, Burn J (1993) Brachmann-de Lange syndrome. Delineation of the clinical phenotype. Am J Med Genet 47:959–964
Jackson L, Kline AD, Barr MA, Koch S (1993) de Lange syndrome: a clinical review of 310 individuals. Am J Med Genet 47:940–946
Jeppsson K, Kanno T, Shirahige K, Sjogren C (2014) The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol 15:601–614
Kagey MH, Newman JJ, Bilodeau S et al (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430–435
Kaur M, Descipio C, Mccallum J et al (2005) Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet A 138:27–31
Kim TH, Abdullaev ZK, Smith AD et al (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128:1231–1245
Kim JH, Rege M, Valeri J et al (2019a) LADL: light-activated dynamic looping for endogenous gene expression control. Nat Methods 16:633–639
Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H (2019b) Human cohesin compacts DNA by loop extrusion. Science 366:1345–1349
Kishi Y, Gotoh Y (2018) Regulation of chromatin structure during neural development. Front Neurosci 12:874
Kline AD, Grados M, Sponseller P et al (2007) Natural history of aging in Cornelia de Lange syndrome. Am J Med Genet C Semin Med Genet 145C:248–260
Kline AD, Krantz ID, Deardorff MA et al (2017) Cornelia de Lange syndrome and molecular implications of the cohesin complex: abstracts from the 7th biennial scientific and educational symposium 2016. Am J Med Genet A 173:1172–1185
Kline AD, Moss JF, Selicorni A et al (2018) Diagnosis and management of Cornelia de Lange syndrome: first international consensus statement. Nat Rev Genet 19:649–666
Krantz ID, Mccallum J, Descipio C et al (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36:631–635
Lieberman-Aiden E, Van Berkum NL, Williams L et al (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293
Maya-Miles D, Andujar E, Perez-Alegre M et al (2019) Crosstalk between chromatin structure, cohesin activity and transcription. Epigenet Chromatin 12:47
Merkenschlager M, Nora EP (2016) CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev Genomics Hum Genet 17:17–43
Miele A, Dekker J (2008) Long-range chromosomal interactions and gene regulation. Mol Biosyst 4:1046–1057
Misteli T (2008) Physiological importance of RNA and protein mobility in the cell nucleus. Histochem Cell Biol 129:5–11
Morgan SL, Mariano NC, Bermudez A et al (2017) Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun 8:15993
Nasmyth K (2011) Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol 13:1170–1177
Nora EP, Lajoie BR, Schulz EG et al (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485:381–385
Nora EP, Goloborodko A, Valton AL et al (2017) Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169(930–944):e22
Ong CT, Corces VG (2014) CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15:234–246
Parelho V, Hadjur S, Spivakov M et al (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422–433
Pauli A, Althoff F, Oliveira RA et al (2008) Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell 14:239–251
Phillips-Cremins JE (2014) Unraveling architecture of the pluripotent genome. Curr Opin Cell Biol 28:96–104
Rajarajan P, Borrman T, Liao W, et al (2018) Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362(6420):eaat4311
Rajarajan P, Borrman T, Liao W et al (2019) Spatial genome exploration in the context of cognitive and neurological disease. Curr Opin Neurobiol 59:112–119
Rao SSP, Huang SC, Glenn St Hilaire B et al (2017) Cohesin loss eliminates all loop domains. Cell 171(305–320):e24
Rowley MJ, Corces VG (2018) Organizational principles of 3D genome architecture. Nat Rev Genet 19:789–800
Rubio ED, Reiss DJ, Welcsh PL et al (2008) CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA 105:8309–8314
Sanborn AL, Rao SS, Huang SC et al (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA 112:E6456–E6465
Sasaki T, Kaga K, Ohira Y, Ogawa Y, Fukushima Y (1996) Temporal bone and brain stem histopathological findings in Cornelia de Lange syndrome. Int J Pediatr Otorhinolaryngol 36:195–204
Schmitt AD, Hu M, Ren B (2016) Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol 17:743–755
Schoenfelder S, Fraser P (2019) Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet 20:437–455
Schuldiner O, Berdnik D, Levy JM et al (2008) piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell 14:227–238
Schwarzer W, Abdennur N, Goloborodko A et al (2017) Two independent modes of chromatin organization revealed by cohesin removal. Nature 551:51–56
Seitan VC, Faure AJ, Zhan Y et al (2013) Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res 23:2066–2077
Sexton T, Cavalli G (2015) The role of chromosome domains in shaping the functional genome. Cell 160:1049–1059
Sofueva S, Yaffe E, Chan WC et al (2013) Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 32:3119–3129
Spielmann M, Lupianez DG, Mundlos S (2018) Structural variation in the 3D genome. Nat Rev Genet 19:453–467
Spitz F (2016) Gene regulation at a distance: from remote enhancers to 3D regulatory ensembles. Semin Cell Dev Biol 57:57–67
Splinter E, Heath H, Kooren J et al (2006) CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 20:2349–2354
Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27:654–666
Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641
Uhlmann F (2016) SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol 17:399–412
Van Den Berg DLC, Azzarelli R, Oishi K et al (2017) Nipbl interacts with Zfp609 and the integrator complex to regulate cortical neuron migration. Neuron 93:348–361
Van Steensel B, Dekker J (2010) Genomics tools for unraveling chromosome architecture. Nat Biotechnol 28:1089–1095
Van Steensel B, Furlong EEM (2019) The role of transcription in shaping the spatial organization of the genome. Nat Rev Mol Cell Biol 20:327–337
Vuilleumier N, Kovari E, Michon A et al (2002) Neuropathological analysis of an adult case of the Cornelia de Lange syndrome. Acta Neuropathol 104:327–332
Watson LA, Wang X, Elbert A, Kernohan KD, Galjart N, Berube NG (2014) Dual effect of CTCF loss on neuroprogenitor differentiation and survival. J Neurosci 34:2860–2870
Wendt KS, Yoshida K, Itoh T et al (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796–801
Whitehead MT, Nagaraj UD, Pearl PL (2015) Neuroimaging features of Cornelia de Lange syndrome. Pediatr Radiol 45:1198–1205
Wittmann M, Queisser G, Eder A et al (2009) Synaptic activity induces dramatic changes in the geometry of the cell nucleus: interplay between nuclear structure, histone H3 phosphorylation, and nuclear calcium signaling. J Neurosci 29:14687–14700
Yamada T, Yang Y, Valnegri P et al (2019) Sensory experience remodels genome architecture in neural circuit to drive motor learning. Nature 569:708–713
Yamaguchi K, Ishitobi F (1999) Brain dysgenesis in Cornelia de Lange syndrome. Clin Neuropathol 18:99–105
Yuan B, Pehlivan D, Karaca E et al (2015) Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Invest 125:636–651
Zhang Y, Mccord RP, Ho YJ et al (2012) Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148:908–921
Zheng H, Xie W (2019) The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 20:535–550
Zuin J, Dixon JR, Van Der Reijden MI et al (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA 111:996–1001
Acknowledgements
The author is grateful to Prof. Toshihide Yamashita (Osaka University) for his support. This work was partly supported by grants from the Japan Society for the Promotion of Science KAKENHI 19K07266), The author received the Encouragement Award of the Japanese Association of Anatomists for fiscal year 2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fujita, Y. Regulation and dysregulation of spatial chromatin structure in the central nervous system. Anat Sci Int 96, 179–186 (2021). https://doi.org/10.1007/s12565-020-00567-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12565-020-00567-7