Abstract
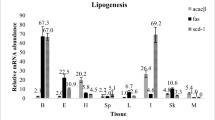
Lipoprotein lipase (LPL) plays a critical role in the metabolism of circulating triacylglycerols (TAGs) packaged in lipoproteins. Fish have two LPL genes, encoding the LPL1 and LPL2 enzymes, but their functions are not yet clearly understood. To provide a basis for further investigation, we cloned a cDNA encoding LPL from the liver of adult medaka Oryzias latipes. The cloned LPL gene was found to share a high identity with LPL1 genes from several fish species and was denoted medaka LPL1. Screening against publicly available medaka expressed sequence tag (EST) databases revealed that LPL1 transcripts are abundant in ESTs from various developmental stages and tissues. The EST screening also identified a second LPL-like gene, denoted medaka LPL2, the transcripts of which were found exclusively in EST libraries from embryonic stages. The LPL1 cDNA obtained by a combination of 3′ and 5′ rapid amplification of cDNA ends was 2,259 bp, with an open reading frame of 1,551 bp encoding 516 amino acids. Quantitative real-time PCR further confirmed the ubiquitous distribution of LPL1 transcripts, with high levels in the liver and visceral adipose tissue, and the lowest level in the intestine. A relatively high expression of the LPL1 gene was also found in the brain, suggesting the potential function of LPL1 in this organ.



Similar content being viewed by others
References
Smith LC, Pownall HJ, Gotto AM Jr (1978) The plasma lipoproteins: structure and metabolism. Annu Rev Biochem 47:751–777
Rogie A, Skinner RE (1985) The roles of the intestine and liver in the biosynthesis of plasma lipoproteins in the rainbow trout, Salmo gairdnerii Richardson. Comp Biochem Physiol B 81:285–289
Nilsson-Ehle P, Garfinkel AS, Schotz MC (1980) Lipolytic enzymes and plasma lipoprotein metabolism. Annu Rev Biochem 49:667–693
Mahley RW, Innerarity TL, Rall SC Jr, Weisgraber KH (1984) Plasma lipoproteins: apolipoprotein structure and function. J Lipid Res 25:1277–1294
Goldberg IJ (1996) Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J Lipid Res 37:693–707
Wang H, Eckel RH (2009) Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab 297:E271–E288
Beigneux AP, Davies BS, Gin P, Weinstein MM, Farber E, Qiao X, Peale F, Bunting S, Walzem RL, Wong JS, Blaner WS, Ding ZM, Melford K, Wongsiriroj N, Shu X, de Sauvage F, Ryan RO, Fong LG, Bensadoun A, Young SG (2007) Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab 5:279–291
Berryman DE, Bensadoun A (1995) Heparan sulfate proteoglycans are primarily responsible for the maintenance of enzyme activity, binding, and degradation of lipoprotein lipase in Chinese hamster ovary cells. J Biol Chem 270:24525–24531
Rumsey SC, Obunike JC, Arad Y, Deckelbaum RJ, Goldberg IJ (1992) Lipoprotein lipase-mediated uptake and degradation of low density lipoproteins by fibroblasts and macrophages. J Clin Invest 90:1504–1512
Fernández-Borja M, Bellido D, Vilella E, Olivecrona G, Vilaró S (1996) Lipoprotein lipase-mediated uptake of lipoprotein in human fibroblasts: evidence for an LDL receptor-independent internalization pathway. J Lipid Res 37:464–481
Weinstock PH, Bisgaier CL, Aalto-Setala K, Radner H, Ramakrishnan R, Levak-Frank S, Essenburg AD, Zechner R, Breslow JL (1995) Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice—mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J Clin Invest 96:2555–2568
Gotoda T, Senda M, Gamou T, Furuichi Y, Oka K (1989) Nucleotide sequence of human cDNA coding for a lipoprotein lipase (LPL) cloned from placental cDNA library. Nucleic Acids Res 17:2351
Semenkovich CF, Chen SH, Wims M, Luo CC, Li WH, Chan L (1989) Lipoprotein lipase and hepatic lipase mRNA tissue specific expression, developmental regulation, and evolution. J Lipid Res 30:423–431
Kirchgessner TG, Svenson KL, Lusis AJ, Schotz MC (1987) The sequence of cDNA encoding lipoprotein lipase. A member of a lipase gene family. J Biol Chem 262:8463–8466
Senda M, Oka K, Brown WV, Qasba PK, Furuichi Y (1987) Molecular cloning and sequence of a cDNA coding for bovine lipoprotein lipase. Proc Natl Acad Sci USA 84:4369–4373
Lindberg A, Olivecrona G (2002) Lipoprotein lipase from rainbow trout differs in several respects from the enzyme in mammals. Gene 292:213–223
Saera-Vila A, Calduch-Giner JA, Gomez-Requeni P, Medale F, Kaushik S, Perez-Sanchez J (2005) Molecular characterization of gilthead sea bream (Sparus aurata) lipoprotein lipase. Transcriptional regulation by season and nutritional condition in skeletal muscle and fat storage tissues. Comp Biochem Physiol B 142:224–232
Merkel M, Eckel RH, Goldberg IJ (2002) Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res 43:1997–2006
Zechner R (1997) The tissue-specific expression of lipoprotein lipase: implications for energy and lipoprotein metabolism. Curr Opin Lipidol 8:77–88
Vilaró S, Llobera M, Bengtsson-Olivecrona G, Olivecrona T (1988) Synthesis of lipoprotein lipase in the liver of newborn rats and localization of the enzyme by immunofluorescence. Biochem J 249:549–556
Gimenez-Llort L, Vilanova J, Skottova N, Bengtsson-Olivecrona G, Llobera M, Robert MQ (1991) Lipoprotein lipase enables triacylglycerol hydrolysis by perfused newborn rat liver. Am J Physiol 261:G641–G647
Cheng HL, Sun SP, Peng YX, Shi XY, Shen X, Meng XP, Dong ZG (2010) cDNA sequence and tissues expression analysis of lipoprotein lipase from common carp (Cyprinus carpio Var. Jian). Mol Biol Rep 37:2665–2673
Cheng HL, Wang X, Peng YX, Meng XP, Sun SP, Shi XY (2009) Molecular cloning and tissue distribution of lipoprotein lipase full-length cDNA from Pengze crucian carp (Carassius auratusvar. Pengze). Comp Biochem Physiol B 153:109–115
Oku H, Koizumi N, Okumura T, Kobayashi T, Umino T (2006) Molecular characterization of lipoprotein lipase, hepatic lipase and pancreatic lipase genes: effects of fasting and refeeding on their gene expression in red sea bream Pagrus major. Comp Biochem Physiol B 145:168–178
Kaneko G, Yamada T, Han Y, Hirano Y, Khieokhajonkhet A, Shirakami H, Nagasaka R, Kondo H, Hirono I, Ushio H, Watabe S (2013) Differences in lipid distribution and expression of peroxisome proliferator-activated receptor gamma and lipoprotein lipase genes in torafugu and red seabream. Gen Comp Endocrinol 184:51–60
Wittbrodt J, Shima A, Schartl M (2002) Medaka—a model organism from the far East. Nat Rev Genet 3:53–64
Kinoshita M, Toyohara H, Sakaguchi M, Inoue K, Yamashita S, Satake M, Wakamatsu Y, Ozato K (1996) A stable line of transgenic medaka (Oryzias latipes) carrying the CAT gene. Aquaculture 143:267–276
Ono Y, Kinoshita S, Ikeda D, Watabe S (2010) Early development of medaka Oryzias latipes muscles as revealed by transgenic approaches using embryonic and larval types of myosin heavy chain genes. Dev Dyn 239:1807–1817
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Zhang ZB, Hu JY (2007) Development and validation of endogenous reference genes for expression profiling of medaka (Oryzias latipes) exposed to endocrine disrupting chemicals by quantitative real-time RT-PCR. Toxicol Sci 95:356–368
Cardin AD, Weintraub HJ (1989) Molecular modeling of protein–glycosaminoglycan interactions. Arteriosclerosis 9:21–32
Dugi KA, Dichek HL, Santamarina-Fojo S (1995) Human hepatic and lipoprotein lipase: the loop covering the catalytic site mediates lipase substrate specificity. J Biol Chem 270:25396–25401
van Tilbeurgh H, Roussel A, Lalouel JM, Cambillau C (1994) Lipoprotein lipase. Molecular model based on the pancreatic lipase x-ray structure: consequences for heparin binding and catalysis. J Biol Chem 269:4626–4633
Wang CS, Hartsuck J, McConathy WJ (1992) Structure and functional properties of lipoprotein lipase. Biochim Biophys Acta 1123:1–17
Wong H, Davis RC, Thuren T, Goers JW, Nikazy J, Waite M, Schotz MC (1994) Lipoprotein lipase domain function. J Biol Chem 269:10319–10323
Henderson HE, Ma Y, Liu MS, Clark-Lewis I, Maeder DL, Kastelein JJ, Brunzell JD, Hayden MR (1993) Structure-function relationships of lipoprotein lipase: mutation analysis and mutagenesis of the loop region. J Lipid Res 34:1593–1602
Wong H, Davis RC, Nikazy J, Seebart KE, Schotz MC (1991) Domain exchange: characterization of a chimeric lipase of hepatic lipase and lipoprotein lipase. Proc Natl Acad Sci USA 88:11290–11294
Hata A, Ridinger DN, Sutherland S, Emi M, Shuhua Z, Myers RL, Ren K, Cheng T, Inoue I, Wilson DE et al (1993) Binding of lipoprotein lipase to heparin. Identification of five critical residues in two distinct segments of the amino-terminal domain. J Biol Chem 268:8447–8457
Ma Y, Henderson HE, Liu MS, Zhang H, Forsythe IJ, Clarke-Lewis I, Hayden MR, Brunzell JD (1994) Mutagenesis in four candidate heparin binding regions (residues 279–282, 291–304, 390–393, and 439–448) and identification of residues affecting heparin binding of human lipoprotein lipase. J Lipid Res 35:2049–2059
Williams SE, Inoue I, Tran H, Fry GL, Pladet MW, Iverius PH, Lalouel JM, Chappell DA, Strickland DK (1994) The carboxyl-terminal domain of lipoprotein lipase binds to the low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor (LRP) and mediates binding of normal very low density lipoproteins to LRP. J Biol Chem 269:8653–8658
Krapp A, Zhang HF, Ginzinger D, Liu MS, Lindberg A, Olivecrona G, Hayden MR, Beisiegel U (1995) Structural features in lipoprotein lipase necessary for the mediation of lipoprotein uptake into cells. J Lipid Res 36:2362–2373
Lo JY, Smith LC, Chan L (1995) Lipoprotein lipase: role of intramolecular disulfide bonds in enzyme catalysis. Biochem Biophys Res Commun 206:266–271
Keiper T, Schneider JG, Dugi KA (2001) Novel site in lipoprotein lipase (LPL415-438) essential for substrate interaction and dimer stability. J Lipid Res 42:1180–1186
Raisonnier A, Etienne J, Arnault F, Brault D, Noe L, Chuat JC, Galibert F (1995) Comparison of the cDNA and amino acid sequences of lipoprotein lipase in eight species. Comp Biochem Physiol B 111:385–398
Bessesen DH, Richards CL, Etienne J, Goers JW, Eckel RH (1993) Spinal cord of the rat contains more lipoprotein lipase than other brain regions. J Lipid Res 34:229–238
Ben-Zeev O, Doolittle MH, Singh N, Chang CH, Schotz MC (1990) Synthesis and regulation of lipoprotein lipase in the hippocampus. J Lipid Res 31:1307–1313
Vilaro S, Camps L, Reina M, Perez-Clausell J, Llobera M, Olivecrona T (1990) Localization of lipoprotein lipase to discrete areas of the guinea pig brain. Brain Res 506:249–253
Gong H, Dong W, Rostad SW, Marcovina SM, Albers JJ, Brunzell JD, Vuletic S (2013) Lipoprotein lipase (LPL) is associated with neurite pathology and its levels are markedly reduced in the dentate gyrus of Alzheimer’s disease brains. J Histochem Cytochem 61:857–868
Koch S, Donarski N, Goetze K, Kreckel M, Stuerenburg HJ, Buhmann C, Beisiegel U (2001) Characterization of four lipoprotein classes in human cerebrospinal fluid. J Lipid Res 42:1143–1151
Brecher P, Kuan HT (1979) Lipoprotein lipase and acid lipase activity in rabbit brain microvessels. J Lipid Res 20:464–471
Wang H, Astarita G, Taussig MD, Bharadwaj KG, DiPatrizio NV, Nave KA, Piomelli D, Goldberg IJ, Eckel RH (2011) Deficiency of lipoprotein lipase in neurons modifies the regulation of energy balance and leads to obesity. Cell Metab 13:105–113
Goti D, Balazs Z, Panzenboeck U, Hrzenjak A, Reicher H, Wagner E, Zechner R, Malle E, Sattler W (2002) Effects of lipoprotein lipase on uptake and transcytosis of low density lipoprotein (LDL) and LDL-associated α-tocopherol in a porcine in vitro blood-brain barrier model. J Biol Chem 277:28537–28544
Xian X, Liu T, Yu J, Wang Y, Miao Y, Zhang J, Yu Y, Ross C, Karasinska JM, Hayden MR, Liu G, Chui D (2009) Presynaptic defects underlying impaired learning and memory function in lipoprotein lipase-deficient mice. J Neurosci 29:4681–4685
Nuñez M, Peinado-Onsurbe J, Vilaró S, Llobera M (1995) Lipoprotein lipase activity in developing rat brain areas. Biol Neonate 68:119–127
Blain JF, Paradis E, Gaudreault SB, Champagne D, Richard D, Poirier J (2004) A role for lipoprotein lipase during synaptic remodeling in the adult mouse brain. Neurobiol Dis 15:510–519
Paradis E, Clement S, Julien P, Ven Murthy MR (2003) Lipoprotein lipase affects the survival and differentiation of neural cells exposed to very low density lipoprotein. J Biol Chem 278:9698–9705
Bundgaard M, Abbott NJ (2008) All vertebrates started out with a glial blood–brain barrier 4–500 million years ago. Glia 56:699–708
Acknowledgments
We acknowledge the National Institute for Basic Biology for the medaka EST data. This work was partly supported by a grant from the Scientific Technique Research Promotion Program for Agriculture, Forestry, Fisheries and Food industry and by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L., Kaneko, G., Takahashi, SI. et al. Identification and gene expression profile analysis of a major type of lipoprotein lipase in adult medaka Oryzias latipes . Fish Sci 81, 163–173 (2015). https://doi.org/10.1007/s12562-014-0826-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-014-0826-7