Abstract
SARS-CoV-2 3C-like protease (3CLpro), a potential therapeutic target for COVID-19, consists of a chymotrypsin fold and a C-terminal α-helical domain (domain III), the latter of which mediates dimerization required for catalytic activation. To gain further understanding of the functional dynamics of SARS-CoV-2 3CLpro, this review extends the scope to the comparative study of many crystal structures of proteases having the chymotrypsin fold (clan PA of the MEROPS database). First, the close correspondence between the zymogen-enzyme transformation in chymotrypsin and the allosteric dimerization activation in SARS-CoV-2 3CLpro is illustrated. Then, it is shown that the 3C-like proteases of family Coronaviridae (the protease family C30), which are closely related to SARS-CoV-2 3CLpro, have the same homodimeric structure and common activation mechanism via domain III mediated dimerization. The survey extended to order Nidovirales reveals that all 3C-like proteases belonging to Nidovirales have domain III, but with various chain lengths, and 3CLpro of family Mesoniviridae (family C107) has the same homodimeric structure as that of C30, even though they have no sequence similarity. As a reference, monomeric 3C proteases belonging to the more distant family Picornaviridae (family C3) lacking domain III are compared with C30, and it is shown that the 3C proteases are rigid enough to maintain their structures in the active state.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The COVID-19 pandemic has had a significant impact on every sector of society worldwide. The extensive response to the pandemic from the biological science community is evident in the enormous number of publications on COVID-19 and SARS-CoV-2. More than 283,000 articles have been collected in the literature hub of COVID-19, LitCovid (08/2022; Chen et al. 2021), since the outbreak. This review focuses on SARS-CoV-2 3C-like protease (3CLpro, also known as main protease), which has been the subject of over 1860 publications. SARS-CoV-2 3CLpro is a cysteine protease that has an important role in viral replication by cleaving the replicase polyprotein to release functional proteins. As such, it is considered a potential target for the development of antiviral therapeutics (Ullrich and Nitsche 2020; Banerjee et al. 2021; Owen et al. 2021; Unoh et al. 2022). Because of the extensive efforts focused on structure-based drug discovery of an inhibitor, more than 580 crystal structures of SARS-CoV-2 3CLpro, most of which are complexed with various drug candidates, have been deposited into the Protein Data Bank (PDB; 08/2022; Berman et al. 2003; Kinjo et al. 2017).
This vast amount of the structural information accumulated in the PDB is an invaluable resource, not only for providing the binding poses of various ligands (Gilson et al. 2016; Wang et al. 2020a, b), but also for constituting a basis for the functional dynamics (Best et al. 2006; Kidera et al. 2021). Interactions with a bound ligand alter the structure of the receptor protein in different ways to produce structural variations in the protein depending on their binding poses (Boehr et al. 2009; Feixas et al. 2014). Considering that crystal packing and amino acid mutations also affect protein structure (Andrec et al. 2007; Cavasotto and Phatak 2009), many crystal structures are affected differently by these factors and constitute a structural ensemble, which is termed “crystal structure ensemble” (Kidera et al. 2021). As the theories of protein dynamics, the conformational selection (Ma et al. 1999) and the linear response theory (Ikeguchi et al. 2005) state that the structural change occurring as a response to external stimulation is a reflection of its intrinsic dynamics, and the crystal structure ensemble can be regarded as a sampled subset of the native structural ensemble (Best et al. 2006; Kidera et al. 2021). Based on the crystal structure ensemble consisting of 343 PDB entries (490 independent chains) of SARS-CoV-2 3CLpro (PDB version 07/21), together with those of highly homologous SARS-CoV 3CLpro (96% identity with SARS-CoV-2 3CLpro; SARS-CoV is the etiological agent of SARS in 2002), we examined the functional dynamics of SARS-CoV-2 3CLpro (Kidera et al. 2021).
In this review, we extend the structural analysis of SARS-CoV-2 3CLpro to the structures of various proteases in clan PA, classified by the MEROPS peptidase database (Rawlings et al. 2018). SARS-CoV-2 3CLpro adopts a highly ubiquitous chymotrypsin fold that is the hallmark of clan PA and belongs to the protease family C30 (Rawlings et al. 2018). As shown in the phylogeny of clan PA (Fig. 1), the chymotrypsin fold ubiquitously appears from animal to virus and has either a serine or cysteine residue as the nucleophile in the catalytic triad/dyad. This versatility of the chymotrypsin fold is a key to understanding the function of SARS-CoV-2 3CLpro and hints at the necessity of comparing SARS-CoV-2 3CLpro with other members of clan PA. Here the crystal structures of the protease families C30, C107, and S32, which belong to order Nidovirales (vertebrate and invertebrate host; Gorbalenya et al. 2006), are summarized in terms of the activating dimerization tightly coupled with the C-terminal α-helical domain III (the crystal structures are listed in Table S1A and the representative structures are shown in Fig. 2). Domain III in C30 of family Coronaviridae has a key role in allosterically activating dimerization (the superimposed structures taken from each species are shown in Fig. S1; Goyal and Goyal 2020). C107 of family Mesoniviridae also forms a homodimer with domain III of the same fold as that of C30 (Fig. 2b), whereas S32 of family Arteriviridae is monomeric with a half-sized C-terminal domain (Fig. 2c and d). Through a comparison of these proteases, we discuss the role of domain III in dimerization. As a reference, we also compared two families of clan PA with no extra C-terminal domain, including family S1 of the animal digestive enzyme and family C3 of cysteine proteases [3C proteases (3Cpro)]. The latter belongs to order Picornavirales (uni- and multi-cellular eukaryote host) whose name 3Cpro is the origin of the name 3CLpro of C30 and is focused on the activation mechanism (the list of the crystal structures are summarized in Table S2; note that 3C indicates the genome position and C3 is the family name in MEROPS). Family S1 is activated by the zymogen-enzyme transformation, which suggests a similarity to the allosteric dimerization activation in C30, whereas monomeric C3 does not have such an activation mechanism. Based on these crystal structures and the relevant literatures, the functional implication of SARS-CoV-2 3CLpro is reviewed. Here, it is noted that both the classification for biological organisms (the taxonomic genera/families) and the classification for molecules (the MEROPS families of proteases) are used concurrently. The former is written in italics, and the latter is designated by the format: “S/C + number” of MEROPS (Rawlings et al. 2018).
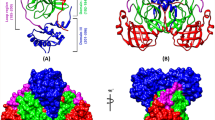
a Taxonomy of viruses belonging to clan PA. The hierarchical classification covers from kingdom/domain/realm to genus, where viruses are under the realm Riboviria. The numbers after the names are the family names of the proteases according to the MEROPS database (Rawlings et al. 2018). In the text, both the virus taxonomy and the protease classification of MEROPS are used. The names in blue are those discussed in this review. The two phyla of virus are positive-sense single-stranded RNA (+ ssRNA) viruses. The members of Nidovirales are given the chain length of 3CLpro listed in MEROPS, along with the variation in each species. Coronaviridae C30 is connected to the detailed phylogenetic tree in (b). At the top of the tree, a cartoon picture of the chymotrypsin fold (chymotrypsinogen, PDB: 1chg) is shown, as the hallmark for clan PA. b Phylogenetic tree of Coronaviridae C30 proteases calculated by the sequences of 3CLpro belonging to Coronaviridae using COBALT (Papadopoulos and Agarwala 2007) and the neighbor joining method. The proteases are those listed in Table S1A whose 3D structures are deposited to the PDB. The subfamilies are designated by the names in blue, Alpha-, Beta-, Gamma-, and Deltacoronavirus
a Homodimer of SARS-CoV-2 3CLpro (PDB: 6m03; family C30). b Homodimer of Cavally virus 3CLpro (5lac; family C107). c Equine arteritis virus 3CLpro (1mbm; family S32). d Porcine reproductive and respiratory syndrome virus 3CL.pro (5y4l; S32). The catalytic dyad for C30 and C107 and the catalytic triad for S32 are drawn in stick. These structures have the chymotrypsin fold (domains I and II) at the N-terminus and domain III at the C-terminus. The size of domain III C30 and C107 is twice that of S32
Catalytic activation allosterically induced by dimerization and ligand binding in SARS-CoV-2 3CLpro
The phylogenetic tree of C30 3CLpro is shown in Fig. 1b. The first crystal structure was solved in 2002, which was 3CLpro of porcine transmissible gastroenteritis coronavirus (TGEV) belonging to genus Alphacoronavirus (PDB:1lvo; Anand et al. 2002). Figure S1 shows the representative structures of the C30 proteases. Anand et al. (2002) revealed that 3CLpro forms a homodimer with the N-terminal Ser1 located near the active site of the partner protomer. These observations were supported by mutational studies of TGEV 3CLpro, in which deletion mutants of domain III (Δ200‒302) and of the five N-terminal residues (Δ1‒5) nearly abolished the proteolytic activity, suggesting that the activation requires domain III and the N-terminal residues (later called the N-finger; Yang et al. 2003), which plays a central role in dimerization (Anand et al. 2002). Shi et al. (2004) confirmed in SARS-CoV 3CLpro that proteolytic activity requires dimerization based on the experiments using dynamic light scattering. Moreover, the importance of the N-finger in dimerization was demonstrated by Hsu et al. (2005a, b) using an ultracentrifuge experiment on SARS-CoV 3CLpro. The importance of domain III to the protease activity was also demonstrated by deletion mutants of human coronavirus 229E 3CLpro (Alphacoronavirus; Ziebuhr et al. 1997), murine coronavirus 3CLpro (Betacoronavirus; Lu and Denison 1997), and avian coronavirus 3CLpro (Gammacoronavirus; Ng and Liu 2000). The importance of the N-finger was also found in porcine epidemic diarrhea virus 3CLpro (Alphacoronavirus; Ye et al. 2016). Therefore, the role of domain III and the N-finger in dimerization and catalytic activity are common to all member of family C30.
The molecular mechanism of the activation became clear when the first crystal structure of SARS-CoV 3CLpro was solved (PDB:1uj1; Yang et al. 2003). The protomers of the dimer in this crystal structure assume different structures (i.e., chain A is in the active state and chain B is in the inactive collapsed state; Fig. 3a), because it is in a highly heterogeneous environment of the crystal in the space group P 1 21 1 (Kidera et al. 2021). For chain A, the inter-protomer hydrogen bond (HB) is formed between Ser1 of chain B and Phe140 of chain A to induce the cooperative formation of many HBs, G138-H172, S139-Y126, L141-Y118, G143-N28, and C145-N28 (Fig. 3a, also see Table S3), which stabilize the active conformation of the residues 138–145 [called the C-loop as it contains the nucleophile Cys145 of the catalytic dyad (Kidera et al. 2021), also called the oxyanion hole loop named after the corresponding loop in lipase (van Tilbeurgh et al. 1993) and the L1 loop by Tan et al. (2005)]. For chain B, the C-loop is collapsed with no HB to the other part of the protein except HB_C145O-N28N (HB between atom O of Cys145 and atom N of Asn28). Of note, the N-finger of chain A is disordered at Ser1 and Gly2 and fails to form the inter-protomer interaction with the C-loop of chain B. HB_C145O-N28N is maintained in both chains, whereas HB_G143O-N28ND2 is only formed in the active chain A (because of this finding, HB_G143O-N28ND2 is regarded as a marker of the active state (Kidera et al. 2021)), resulting in the formation/collapse transition of the oxyanion hole (Fig. 3a; the NH groups of Gly143 and Cys145 together stabilize the oxyanion at Gln in the substrate, which occurs as a reaction intermediate of the proteolytic reaction; Otto and Schirmeister 1997). Clear evidence of the coupling between the proteolytic activity and dimerization was presented in the monomeric crystal structures of dimerization-deficient mutants of SARS-CoV 3CLpro [PDB:2pwx (G11A), 2qcy (R298A), 3f9e (S139A), and 3m3t (R298A); Chen et al. 2008; Shi et al. 2008; Hu et al. 2009]. These monomeric structures contain a collapsed C-loop and lose all HBs listed above, except for HB_C145O-N28N. The cooperative structural transition of the C-loop allosterically triggered by HB_F140O-S1N* (* indicates the other protomer) was shown to be a common feature in SARS-CoV-2 and SARS-CoV 3CLpro (Kidera et al. 2021).
a The structure of the C-loop in SARS-CoV 3CLpro. Active state (left; PDB: 1uj1_A) and collapsed inactive state (right; 1uj1_B). The active state is defined by the presence of the oxyanion hole (the main-chain NH groups of Gly143 and Cys145 are ready to catch an oxyanion as the reaction intermediate of the substrate). This is monitored by the main-chain oxygen atoms of Gly143 and Cys145, both of which have HBs with Asn28 in the active state and HB_G143-N28 is lost in the collapsed state. Since HB_C145-N28 is maintained even in the collapsed state, the main-chain NH group of Gly143 goes down to the N-terminal side of the C-loop, indicated by the broken lines, to collapse the oxyanion hole. b The structure of the C-loop in chymotrypsin/chymotrypsinogen. Active state (left; PDB: 4cha, chymotrypsin) and collapsed state (right; 1ex3, chymotrypsinogen). Zymogen activation causes a cleavage at Ile16 to produce the positive charge at the N-terminus. The charged Ile16 forms a salt bridge with the carboxyl group of Asp194. In the zymogen state, the side-chain Asp194 forms a polar contact with His40, and salt bridge formation in the activation causes a large flip of the side-chain of Asp194 to make the conformational change in the C-loop to form HB_G193_H40. This bond enables Gly193 and Cys195 to form the oxyanion hole
Another factor that contributes to the activation of 3CLpro is substrate/ligand binding. Cheng et al. (2010) demonstrated substrate-induced dimerization by showing that the dimerization-deficient mutant R298A/L of SARS-CoV 3CLpro shifts the equilibrium to the dimer side when the concentration of the substrate peptide is increased. The crystal structure of the R298A mutant of SARS-CoV 3CLpro (PDB:4hi3; Wu et al. 2013) crystalized with a high concentration of a substrate molecule shows a homodimeric structure, in which the C-loop recovered the active state including HB_F140O-S1N*, although the electron density of the substrate was not observed. The recovery of the active C-loop is clearly explained by substrate recognition at the subsite S1 including the oxyanion hole defined by Schechter and Berger (1967). The stabilization of the dimeric form by substrate binding is not straightforward, because the substrate does not directly contribute to the inter-protomer interactions. The following scenario may be considered: the bound substrate induces the active C-loop, which in turn enables HB_F140O-S1N* to form. This HB then sets the proper position of the two protomers to trigger the formation of the other inter-protomer interactions. Many cases of ligand-induced activation were found in the crystal structure ensemble (Kidera et al. 2021). Some amino acids appended to the N-terminus eliminate the positive charge at the N-terminal Ser1 to weaken the interaction with Phe140, although these crystal structures maintain the homodimeric structure (Lee et al. 2005; Xue et al. 2007). As a result, the C-loop conformation tends to be collapsed in the ligand-free state with a frequency of 13/25 (there are 25 ligand-free chains in 3CLpro having some amino acids appended to the N-terminus. Of these, 13 chains have a collapsed C-loop conformation); however, this frequency decreases almost ten-fold to 7/91 in the ligand-bound structures (Kidera et al. 2021). The influence of ligand interactions is a clue to understanding the maturation process of 3CLpro in the replicase polyprotein, in which 3CLpro in a polyprotein, having the extension at both termini, autoclaves itself with the aid of ligand-induced activation to produce the mature 3CLpro (Hsu et al. 2005a, b; Li et al. 2010; Xia and Kang 2011).
Comparison of SARS-CoV-2 3CLpro with family S1: activation and ligand interactions
The zymogen-enzyme transformation is the activation mechanism of S1 proteases (Stroud et al. 1977; S1 is the largest family of proteases, and its nucleophile in the catalytic triad is serine, including trypsin and chymotrypsin). Figure 3b shows a representative case in which the active form of the C-loop of α-chymotrypsin (residues 188–195 containing the nucleophile Ser195 of the catalytic triad; PDB:4cha; Tsukada and Blow 1985) is compared with the inactive form of chymotrypsinogen (the zymogen of chymotrypsin; PDB:1ex3; Pjura et al. 2000). Because chymotrypsinogen is cleaved into three chains, residues 1‒13, 16‒146, and 149‒149, for conversion into α-chymotrypsin, the N-terminal positive charge NH3+ of Ile16 forms a salt bridge with the carboxylate of Asp194 and induces a flipping motion of the Asp194 side-chain from the position hydrogen bonded to His40 in chymotrypsinogen. The dissociation between Asp194 and His40 allows the oxygen atom of Gly193 to form an HB to His40 and to complete the oxyanion hole consisting of the nitrogen atoms of Gly193 and Ser195, whereas Ser195 stably maintains an HB with Gly43 during the whole transformation process (Fig. 3b). The active form has a number of other HBs to the C-loop (S189-V17, M192-L143, D194-G142, and S195-H57, where His57 is the base of the catalytic triad). Another unique point is the disulfide bridge between Cys191 and Cys220, which fixes the N-terminal position of the C-loop, although this disulfide bridge occurs only in the subfamily S1A (family S1 contains the subfamilies S1A-S1F in MEROPS (Rawlings et al. 2018)).
Comparing the conformational changes occurring during the activation process between SARS-CoV 3CLpro and chymotrypsinogen/chymotrypsin, we noticed similarities between the two proteases including the salt bridge with the N-terminal residue as the trigger for activation (Ser1 in another protomer of 3CLpro vs. Ile16 of the cleaved N-terminus in chymotrypsin), the stable position of the catalytic residue before and after activation (HB_C145O-N28N vs. HB_S195O-G43N), the switchable HB of the glycine residue constituting the oxyanion hole (HB_G143O-N28ND2 vs. HB_G193O-H40NE2), and many HBs cooperatively formed to stabilize the active form of the C-loop. Because there is no significant sequence similarity between the two proteases, the similarity in the activation mechanism should be the consequence of the strong evolutional constraint under the environment of the same chymotrypsin fold. However, the implication of the similar regulation for the catalytic activity of SARS-CoV-2 3CLpro remains obscure, whereas the necessity of the strict regulation for the cleavage activity in chymotrypsin is well understood (Stroud et al. 1977).
In addition to the structural changes during activation, another important functional dynamics upon ligand binding occurs in the β-turn at residues 166–178 in SARS-CoV-2 3CLpro, called the E-loop (named after the N-terminal residue of Glu166 at the subsite S3, also called L2 by Tan et al. (2005)). According to the crystal structure ensemble of 3CLpro, the E-loop sensitively responds to the ligand interactions to cause a ligand size-dependent conformational change. A large ligand shifts down the E-loop to make a larger space for the ligand, whereas a small ligand shifts up the E-loop to maintain the interactions (Kidera et al. 2021). The simulation of the binding process of a substrate peptide indicated that the dynamics of the E-loop play an important role in leading the peptide from the initially encountered position at the protein surface to the fully bound state deep inside of the cleft by susceptibly changing its conformation (Moritsugu et al. 2022).
With respect to family S1, thrombin shows a highly dynamic structure at a β-loop-β segment (residues 213–226; known as the Na+ loop containing the Na+ binding sites, Arg221a and Lys224) corresponding to the E-loop in SARS-CoV-2 3CLpro (Gohara and Di Cera 2011; Lechtenberg et al. 2012). Within the fluctuation range, it contains the two conformational states of the slow and fast forms (also called the E* and E forms), which regulate the catalytic activity responding to the inter-molecular interactions including Na+ binding. This dynamic feature in this segment is believed to be shared by various members of family S1 (Gohara and Di Cera 2011; Lechtenberg et al. 2012). A functionally relevant dynamics is also found in the high temperature requirement A (HtrA) proteases, in which the loop corresponding to the E-loop of SARS-CoV-2 3CLpro allosterically changes conformation in the trimeric structure to induce the active structure upon binding of a signal peptide to the C-terminal PDZ domain (Wilken et al. 2004; Krojer et al. 2010; Sawa et al. 2011). These coincidences between the E-loop of SARS-CoV-2 3CLpro and the corresponding loop of the members of family S1 are also considered to be resulted from the strong evolutionary constraint of the chymotrypsin fold.
SARS-CoV-2 3CL.pro within family C30 (family Coronaviridae)
SARS-CoV-2 3CLpro belongs to the protease family C30 (Rawlings et al. 2018), which consists of the four genera of family Coronaviridae, Alpha-, Beta-, Gamma-, and Deltacoronavirus (Woo et al. 2010). In Fig. 1b, the phylogenetic tree of C30 3CLpro was constructed for the species having the PDB entries listed in Table S1A. The classification of 3CLpro is consistent with that of the four genera and the subgenera of viruses defined by the whole genome. SARS-CoV-2 3CLpro is correctly classified in Betacoronavirus. The representative structures of C30 are shown in Fig. S1a after superimposition at the chymotrypsin fold of chain A (N-terminal domains I and II), because domain III and chain B fluctuate largely and randomly against domains I and II of chain A, which primarily depends on the crystal environment. These structures clearly show that the C30 proteases share the same homodimeric form as that of SARS-CoV-2 3CLpro. The significant structural variation in the chymotrypsin fold is found in the E-loop, of which dynamics is relevant to the functional motion in SARS-CoV-2 3CLpro as described above. The other loops having large fluctuations (residues 46–49 and 70–73) are caused mostly by amino acid insertion/deletion. When the C-terminal domain III is superimposed (Fig. S1b), it is found that all 3CLpro have the same fold with large fluctuations at the loop regions (residues 214‒215 and 243‒247), which are also caused by amino acid insertion/deletion.
More detailed structural comparisons were done separately for N-terminal domains I and II and for domain III. The structures of family C30 were subjected to hierarchical clustering based on the Cα RMSD after structural alignment by the CE algorithm (Shindyalov and Bourne 1998). The structural classification of domains I and II (Fig. S2a) is mostly consistent with the sequence classification shown in Fig. S2c, in which the four genera and their subgenera are separately clustered. In contrast in the comparison of domain III, Fig. S2b shows that SARS-CoV-2 is classified outside of the cluster of Betacoronavirus because more significant mutations occurred in domain III of SARS-CoV-2 3CLpro compared with the other members (Fig. S2d). This is more clearly shown in the plot of the percentage of identical amino acids in domain III versus that of domains I and II (Fig. S2e). This figure also shows that 70% of the alignments revealed that domain III accumulates more mutations compared with domains I and II, particularly in SARS-CoV-2. This is because domain III has a larger mutational space under the weaker evolutional constraint probably because of the α-helical structure and the distant location from the active site.
We examined the HB pattern of the C-loop in 3CLpro of family C30, the loop containing the oxyanion hole regulating the catalytic activity. For SARS-CoV-2 3CLpro, the five HBs are cooperatively formed between the main-chain atoms of the C-loop and the side-chain atoms of the surrounding residues except for HB_F140O-S1N*, when allosterically induced by dimerization. In contrast, HB_C145O-N28N is kept both in the active and collapsed states. This pattern of the HBs between the main-chain and side-chain atoms indicates that the transition between the active and the collapsed states occurs exclusively in the C-loop, but the main-chain of the surrounding residues stays in almost the same position throughout the transition, except for the N-finger. Table S3 shows the HB patterns of the C-loop for the members of C30 listed in Table S1A. The same behaviors as those of SARS-CoV-2 3CLpro were observed for the members of C30. The main-chain atoms of the C-loop bind to the side-chain atoms of the surrounding residues in the active state. The transition between the active and collapsed forms is evident in the comparison between the monomeric collapsed form of 2q6d and the ligand-bound dimer of 2q6f (3CLpro of avian infectious bronchitis virus (IBV) belonging to Gammacoronavirus) (Xue et al. 2008). The former has a collapsed C-loop with none of the five HBs formed, whereas the latter is in the active state with all five HBs formed. However, in the other coronavirus 3C-like proteases, there is a marked distinction from SARS-CoV-2 3CLpro, that is, these proteases have, at most, four HBs instead of five except for porcine epidemic diarrhea virus 3CLpro having five HBs. The decrease in the number of the HBs is resulted from the mutation from Tyr to Phe disabling HB formation at the side-chain (Phe125 in human coronavirus 229E, human coronavirus NL63, and Phe129 in Betacoronaviruses other than SARS-CoV-2) and the mutation from Ala/Ser to Thr at position 143, which causes a steric hindrance at the side-chain methyl group to disrupt HB_I140N-Y117OH (porcine transmissible gastroenteritis coronavirus 3CLpro and feline coronavirus 3CLpro). The reason why the active form is maintained in these proteases despite the smaller number of HBs is because ligand binding assists to stabilize the active form of the C-loop, that is, the ligand-induced activation occurs. Tomar et al. (2015) found that MERS-CoV is a weakly associated dimer requiring ligand binding for activation. It is also notable that the PDB entry of 2q6d mentioned above has a monomeric protein in the asymmetric unit, suggesting that IBV 3CLpro has a large population of the monomeric protein even at high concentration during crystallization (Xue et al. 2008). The presence of ligand-induced activation can also be postulated based on the fact that 70% of the PDB entries in Table S1A are in the ligand-bound state.
SARS-CoV-2 3CLpro as a member of Nidovirales: domain III
As shown in Fig. 1a, SARS-CoV-2 belongs to order Nidovirales, whose crystal structures of 3CLpro in the families S32 (PDB:1mbm; Barrette-Ng et al. 2002, 3fan, 3fao; Tian et al. 2009, and 5y4l; Shi et al. 2018), C30 (Table S1A), and C107 (5lac and 5lak; Kanitz et al. 2019) contain the C-terminal domain III in addition to the chymotrypsin fold of domains I and II (Fig. 2). The presence of domain III was also reported in families C62, S65, and S75 based on the sequence analysis (Ziebuhr et al. 2003; Smits et al. 2006; Ulferts et al. 2011). Therefore, the C-terminal domain III is a conserved feature of Nidovirales 3CLpro (Gorbalenya et al. 2006).
The most striking finding regarding Nidovirales is that C107 of family Mesoniviridae, infecting mosquito, is a homodimer of which fold architecture is essentially the same as that of C30 (family Coronaviridae) as shown in Fig. 2b, although there is no significant sequence similarity between the two families (Kanitz et al. 2019). However, a close examination shows that these distantly related proteases exhibit significant structural variations caused by insertions and deletions (Fig. S3). Particularly in domain III, the structural correspondence is not easy to trace. The HB pattern of the C-loop listed in Table S3 indicates that C107 has fewer HBs stabilizing the C-loop, and both of the ligand-free and ligand-bound structures (PDB: 5lac and 5lak, respectively; Kanitz et al. 2019) are in the active state according to the HB pattern for the residues contributing to the oxyanion hole (HB_G151O-R35N and HB_G153O-R35NH1), although neither of them has an HB with S1 of the partner protomer. These observations suggest that the role of HBs is different from that of C30, although the two structures may not be sufficient to draw a definitive conclusion.
Family S32 is monomeric (Fig. 2c and d) and has domain III of half the size of domain III in families C30 and C107. The HB pattern at the oxyanion hole (HB_S120O-T22N (or HB_S118O-S22N) and HB_G118O-T22OG1 (or HB_G116O-S22OG)) shown in Table S3 indicates that the two structures are in the active state (PDB: 1mbm; Barrette-Ng et al. 2002 and 5y4l; Shi et al. 2018), whereas the other two are collapsed (3fan and 3fao; Tian et al. 2009). This suggests that S32 retains the activating transition of the C-loop as is the case for the dimeric C30. This is a marked difference from the monomeric 3Cpro of family C3 (Picornaviridae) discussed below.
To discuss the structures of the members with unknown 3D structures, families C62, S65, and S75, the families in order Nidovirales are roughly classified into two groups: (I) S32, S65, and S75 and (II) C30, C107, and C62, based on the following three pieces of information: (1) The phylogenetic tree of Nidovirales classifies the families into these two groups (Fig. 3 in Gulyaeva and Gorbalenya (2021)); (2) the nucleophile of the proteolytic reaction separates them into the two groups of (I) serine and (II) cysteine groups; (3) domain III has the three different sizes including small (S32), medium (S65 and S75), and large (C30, C107 and C65) as indicated in Fig. 1a. Based on this classification, it is hypothesized that S65 and S75 are monomeric like S32. Xu et al. (2020) successfully built a homology model of S65 using the crystal structure of S32 (PDB: 1mbm) as a template. C62 can be classified into the dimeric C30 and C107 groups, although there is no literature that discusses whether C62 is a homodimer or not.
We further applied AlphaFold 2 and AlphaFold-Multimer (Jumper et al. 2021; Evans et al. 2021) to the structural prediction of the representative members of families C62, S65, and S75. Fig. S4 shows the prediction results. When the predicted monomer structures are compared with the structure of SARS-CoV-2 3CLpro (PDB: 6m03), we found that the chymotrypsin fold are correctly predicted together with the positions of the catalytic residues as indicated by the high confidence level (pLDDT, predicted Local Distance Difference Test) despite no significant sequence similarity between these families and the other members of clan PA. These results indicate that the machine learning of the chymotrypsin fold is at a high level due to the unique 3D structure with the diverse sequences and enables us to predict the chymotrypsin fold for any member of clan PA with high precision. On the other hand, the prediction of domain III was not satisfactory as shown in the right figures and the low confidence level (Fig. S4a-c). Domain III has a unique α-helical fold (InterPro: Peptidase_C30_dom3_CoV; URL: www.ebi.ac.uk/interpro/) and is found only in the limited protease families of C30 and C107. These conditions made the prediction of domain III difficult when there is no significant sequence similarity with C30 or C107.
In the prediction of the homodimer, only family S75 gave a significant result with the low error level (PAE, predicted aligned error) of the inter-protomer arrangement, whereas families C62 and S65 did not provide a reasonable prediction (data not shown). Unexpectedly, the prediction of S75 closely resembles the dimeric structure of C30 (Fig. S4d) despite the low confidence level in domain III. These prediction results appear to suggest that S75 is dimeric, whereas C62 and S65 are monomeric, which are incompatible with the expectation in the above classification. The prediction of the homodimer structures was repeated with the sequences with domain III removed, and basically the same results were obtained. This indicates that the interface between the two chymotrypsin folds is the determinant in the dimer prediction. However, we need a further study on the structures of C62, S65, and S75 to draw a definitive conclusion.
Finally, the role of domain III is discussed by quoting the work of van Aken et al. (2006). They carried out a mutagenesis study on equine arteritis virus 3CLpro (the same protein as PDB:1mbm belonging to family S32) to identify the role of domain III. They demonstrated the importance of not only domain III but also the linker connecting domains II and III (they call it the hinge region) in the proteolytic processing of the replicase polyprotein using mutants of the linker as well as a deletion mutant of domain III. The linker binds the N-terminal part of the substrate by adaptively changing the conformation, whereas domain III does not have a direct interaction with the substrate. Therefore, van Aken et al. (2006) concluded that the linker has an important role in the proteolytic reaction, and domain III works to situate the linker at an appropriate position for catalysis. This point is also argued by Anand et al. (2002). This speculation is supported by the comparison between family C30 containing domain III and family C3 lacking domain III. Figure S5 compares SARS-CoV 3CLpro (C30; PDB: 2q6g, Xue et al. 2008) with coxsackievirus A16 3Cpro, a representative member of C3 belonging to family Picornaviridae (PDB: 3sj9; Lu et al. 2011; also see Fig. 1a). The recognition site at the linker in SARS-CoV 3CLpro is replaced by the elongated loop located at the N-terminus to the C-loop (known as the β-ribbon; Sweeney et al. 2007) in coxsackievirus A16 3Cpro. The β-ribbon of C3 can be stably maintained by itself, whereas the flexible linker of C30 is regulated by domain III. This difference reflects in the cleavage site specificity. According to the substrate specificity data in MEROPS (Rawlings et al. 2018) where the substrate sequences are compiled, the amino acid preference at the peptide site P4 (the substrate amino acid position corresponding to the subsite S4; Schechter and Berger 1967) recognized by the β-ribbon in C3 is more specific compared with the P4 site recognized by the linker in C30. The most predominant amino acid appearing at site P4 accounts for 60% (35/58) of the total amino acid occurrence observed in C3, whereas in C30, it accounts for only up to 36% (43/119) (Rawlings et al. 2018). This suggests that the flexible linker of C30 recognizes a larger variety of amino acids at the P4 site compared with the rigid β-ribbon of C3. However, the linker in C30 is not freely fluctuating, but dimerization restricts the conformational freedom of the linker to a certain level and enables it to susceptibly respond to various substrates.
3C proteases of family C3 (family Picornaviridae)
Family C3, 3Cpro of family Picornaviridae belonging to order Picornavirales (Fig. 1), has the chymotrypsin fold but lacks domain III; thus, it is monomeric (Fig. S6; Sun et al. 2016; Yi et al. 2021). Although 3CLpro of C30 is named after 3Cpro of C3, these two families belong to different orders and have no sequence similarity. Unlike family S1 or C30, which are under allosteric regulation of the catalytic activation, in C3 such regulation is not known. Therefore, C3 likely has a different HB pattern in the C-loop compared with that of C30.
Table S4 summarizes the HB pattern of the C-loop in the protease family C3. Compared with the HB pattern in C30 shown in Table S3, the number of HBs is smaller in C3. The HBs are simply classified into either of the following two types. The first are HBs between the C-loop and the E-loop (HB pairs 1 and 2 in Table S4), which are stably formed between the main-chain atoms even in the collapsed C-loop (PDB: 3zz4 and 3osy; Cui et al. 2011). These HBs are considered to increase the rigidity of the E-loop in C3 compared with the flexible E-loop in C30 (Fig. S1). Recalling the discussion in the previous section that the β-ribbon in C3 was more rigid compared with the linker in C30, we conclude that monomeric C3 is more rigid. The second type of HBs is associated with the main-chain oxygen atoms in the oxyanion hole-forming residues (HB pair 3 and 4 in Table S4). These HBs in C3 exhibit various patterns for each protease subfamily. Subfamily C3E has the same HB pattern as C30 (i.e., HB_C172O-N30N and HB_C170O-N30ND2). Subfamily C3C has HB_C163O-C32N and HB_C161O-N121ND2, in which the HB is formed, not with Cys32, but with Asn121 in the β-turn located on the N-terminus to the β-ribbon, because the side-chain of Cys32 does not provide a hydrogen donor. Subfamily C3A has an HB between Cys147 and Thr26 in only a half of the entries, and the HB to Gly145 is scarcely formed because the hydroxyl group of Thr26 is not strong enough to compete with water for HB formation. Subfamily C3B does not contain these HBs simply because it lacks the secondary structure to generate the HB to the catalytic cysteine residue (Fig. S6). Even though these various HB patterns exist, there are only two entries exhibiting a completely collapsed C-loop (PDB: 3zz4 and 3osy; in 3osy, the β-ribbon in another molecule in the crystal takes the open form to interact with the C-loop and to collapse it). Therefore, these HBs are not required to maintain the C-loop conformation in the active state. These observations suggest that C3 is rigid enough to constantly maintain the structure in the active state.
To summarize the survey on the proteases in clan PA, we found various types of common features originating from the chymotrypsin fold, whereas the features specific to SARS-CoV-2 3CLpro were also found. Among the common features, the ligand molecules bound by proteases of multiple species/families should be remarked here. Tables S1A, B, and S2 cite the ligand names in the crystal structures of family C30 and C3, respectively, where we marked the names when their ligands are shared with SARS-CoV-2/SARS-CoV 3CLpro. The result indicates that the majority of the PDB entries contain ligand molecules which also appear in the crystal structures of SARS-CoV-2/SARS-CoV 3CLpro: 17 entries out of 26 entries in family C30 and 29 entries out of 56 entries in family C3A share the same ligand with the entries of SARS-CoV-2/SARS-CoV 3CLpro. This finding suggests not only the 3D structural similarity of the ligand binding site within the members of clan PA, but also possible contributions of these evolutional relation to the drug discovery problem for SARS-CoV-2 3CLpro and to the development of broad-spectrum inhibitors covering various species and variants belonging to clan PA (Wang et al. 2020a, b; Jukič et al. 2021; Luttens et al. 2022; Ullrich et al. 2022; Uraki et al. 2022).
Concerning the feature specific to SARS-CoV-2 3CLpro, we hypothesize that SARS-CoV-2 3CLpro utilizes the machinery available in the chymotrypsin fold and domain III optimally to achieve the most susceptible regulation of proteolytic function compared with other proteases in clan PA. However, we have not understood the details how and why this susceptible regulation is employed in the proteolytic processing of the replicase polyprotein or in the cleavage of host proteins.
References
Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R (2002) Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J 21:3213–3224. https://doi.org/10.1093/emboj/cdf327
Andrec M, Snyder DA, Zhou Z, Young J, Montelione GT, Levy RM (2007) A large data set comparison of protein structures determined by crystallography and NMR statistical test for structural differences and the effect of crystal packing. Proteins 69:449–465. https://doi.org/10.1002/prot.21507
Banerjee R, Perera L, Tillekeratne LMV (2021) Potential SARS-CoV-2 main protease inhibitors. Drug Discov Today 26:804–816. https://doi.org/10.1016/j.drudis.2020.12.005
Barrette-Ng IH, Ng KK, Mark BL, Van Aken D, Cherney MM, Garen C, Kolodenko Y, Gorbalenya AE, Snijder EJ, James MN (2002) Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J Biol Chem 277:39960–39966. https://doi.org/10.1074/jbc.M206978200
Berman H, Henrick K, Nakamura H (2003) Announcing the worldwide Protein Data Bank. Nat Struct Biol 10:980. https://doi.org/10.1038/nsb1203-980
Best RB, Lindorff-Larsen K, DePristo MA, Vendruscolo M (2006) Relation between native ensembles and experimental structures of proteins. Proc Natl Acad Sci USA 103:10901–10906. https://doi.org/10.1073/pnas.0511156103
Boehr DD, Nussinov R, Wright PE (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5:789–796. https://doi.org/10.1038/nchembio.232
Cavasotto CN, Phatak SS (2009) Homology modeling in drug discovery current trends and applications. Drug Discov Today 14:676–683. https://doi.org/10.1016/j.drudis.2009.04.006
Chen S, Hu T, Zhang J, Chen J, Chen K, Ding J, Jiang H, Shen X (2008) Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease crystal structure with molecular dynamics simulations. J Biol Chem 283:554–564. https://doi.org/10.1074/jbc.M705240200
Chen Q, Allot A, Lu Z (2021) LitCovid: an open database of COVID-19 literature. Nucleic Acids Res 49:D1534–D1540. https://doi.org/10.1093/nar/gkaa952
Cheng SC, Chang GG, Chou CY (2010) Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys J 98:1327–1336. https://doi.org/10.1016/j.bpj.2009.12.4272
Cui S, Wang J, Fan T, Qin B, Guo L, Lei X, Wang J, Wang M, Jin Q (2011) Crystal structure of human enterovirus 71 3C protease. J Mol Biol 408:449–461. https://doi.org/10.1016/j.jmb.2011.03.007
Evans R, O'Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, Ronneberger O, Bodenstein S. Zielinski M. Bridgland A, Potapenko A, Cowie A, Tunyasuvunakool K, Jain R, Clancy E, Hassabis D (2021) Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021.10.04.463034. https://doi.org/10.1101/2021.10.04.463034.
Feixas F, Lindert S, Sinko W, McCammon JA (2014) Exploring the role of receptor flexibility in structure-based drug discovery. Biophys Chem 186:31–45. https://doi.org/10.1016/j.bpc.2013.10.007
Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 44:D1045–D1053. https://doi.org/10.1093/nar/gkv1072
Gohara DW, Di Cera E (2011) Allostery in trypsin-like proteases suggests new therapeutic strategies. Trends Biotechnol 29:577–585. https://doi.org/10.1016/j.tibtech.2011.06.001
Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ (2006) Nidovirales evolving the largest RNA virus genome. Virus Res 117:17–37. https://doi.org/10.1016/j.virusres.2006.01.017
Goyal B, Goyal D (2020) Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb Sci 22:297–305. https://doi.org/10.1021/acscombsci.0c00058
Gulyaeva AA, Gorbalenya AE (2021) A nidovirus perspective on SARS-CoV-2. Biochem Biophys Res Commun 538:24–34. https://doi.org/10.1016/j.bbrc.2020.11.015
Hsu MF, Kuo CJ, Chang KT, Chang HC, Chou CC, Ko TP, Shr HL, Chang GG, Wang AH, Liang PH (2005) Mechanism of the maturation process of SARS-CoV 3CL protease. J Biol Chem 280:31257–31266. https://doi.org/10.1074/jbc.M502577200
Hsu WC, Chang HC, Chou CY, Tsai PJ, Lin PI, Chang GG (2005) Critical assessment of important regions in the subunit association and catalytic action of the severe acute respiratory syndrome coronavirus main protease. J Biol Chem 280:22741–22748. https://doi.org/10.1074/jbc.M502556200
Hu T, Zhang Y, Li L, Wang K, Chen S, Chen J, Ding J, Jiang H, Shen X (2009) Two adjacent mutations on the dimer interface of SARS coronavirus 3C-like protease cause different conformational changes in crystal structure. Virology 388:324–334. https://doi.org/10.1016/j.virol.2009.03.034
Ikeguchi M, Ueno J, Sato M, Kidera A (2005) Protein structural change upon ligand binding: linear response theory. Phys Rev Lett 94:078102. https://doi.org/10.1103/PhysRevLett.94.078102
Jukič M, Škrlj B, Tomšič G, Pleško S, Podlipnik Č, Bren U (2021) Prioritisation of compounds for 3CLpro inhibitor development on SARS-CoV-2 variants. Molecules 26:3003. https://doi.org/10.3390/molecules26103003
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Kanitz M, Blanck S, Heine A, Gulyaeva AA, Gorbalenya AE, Ziebuhr J, Diederich WE (2019) Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus. Virology 533:21–33. https://doi.org/10.1016/j.virol.2019.05.001
Kidera A, Moritsugu K, Ekimoto T, Ikeguchi M (2021) Allosteric regulation of 3CL protease of SARS-CoV-2 and SARS-CoV observed in the crystal structure ensemble. J Mol Biol 433:167324. https://doi.org/10.1016/j.jmb.2021.167324
Kinjo AR, Bekker GJ, Suzuki H, Tsuchiya Y, Kawabata T, Ikegawa Y, Nakamura H (2017) Protein Data Bank Japan (PDBj) updated user interfaces, resource description framework, analysis tools for large structures. Nucleic Acids Res 45:D282–D288. https://doi.org/10.1093/nar/gkw962
Krojer T, Sawa J, Huber R (2010) Clausen T (2010) HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues. Nat Struct Mol Biol 17(7):844–852. https://doi.org/10.1038/nsmb.1840
Lechtenberg BC, Freund SM, Huntington JA (2012) An ensemble view of thrombin allostery. Biol Chem 393:889–898. https://doi.org/10.1515/hsz-2012-0178
Lee TW, Cherney MM, Huitema C, Liu J, James KE, Powers JC, Eltis LD, James MN (2005) Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate-like aza-peptide epoxide. J Mol Biol 353:1137–1151. https://doi.org/10.1016/j.jmb.2005.09.004
Li C, Qi Y, Teng X, Yang Z, Wei P, Zhang C, Tan L, Zhou L, Liu Y, Lai L (2010) Maturation mechanism of severe acute respiratory syndrome (SARS) coronavirus 3C-like proteinase. J Biol Chem 285:28134–28140. https://doi.org/10.1074/jbc.M109.095851
Lu Y, Denison MR (1997) Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology 230:335–342. https://doi.org/10.1006/viro.1997.8479
Lu G, Qi J, Chen Z, Xu X, Gao F, Lin D, Qian W, Liu H, Jiang H, Yan J, Gao GF (2011) Enterovirus 71 and coxsackievirus A16 3C proteasesbinding to rupintrivir and their substrates and anti-hand, foot, and mouth disease virus drug design. J Virol 85:10319–10331. https://doi.org/10.1128/JVI.00787-11
Luttens A, Gullberg H, Abdurakhmanov E, Vo DD, Akaberi D, Talibov VO, Nekhotiaeva N, Vangeel L, De Jonghe S, Jochmans D, Krambrich J, Tas A, Lundgren B, Gravenfors Y, Craig AJ, Atilaw Y, Sandström A, Moodie LWK, Lundkvist Å, van Hemert MJ, Neyts J, Lennerstrand J, Kihlberg J, Sandberg K, Danielson UH, Carlsson J (2022) Ultralarge virtual screening identifies SARS-CoV-2 main protease inhibitors with broad-spectrum activity against coronaviruses. J Am Chem Soc 144:2905–2920. https://doi.org/10.1021/jacs.1c08402
Ma B, Kumar S, Tsai CJ, Nussinov R (1999) Folding funnels and binding mechanisms. Protein Eng 12:713–720. https://doi.org/10.1093/protein/12.9.713
Moritsugu K, Ekimoto T, Ikeguchi M, Kidera A (2022) Binding and unbinding pathways of peptide substrate on SARS-CoV-2 3CL protease. bioRxiv 2022.06.08.495396. https://doi.org/10.1101/2022.06.08.495396.
Ng LF, Liu DX (2000) Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology 272:27–39. https://doi.org/10.1006/viro.2000.0330
Otto HH, Schirmeister T (1997) Cysteine proteases and their inhibitors. Chem Rev 97:133–172. https://doi.org/10.1021/cr950025u
Owen DR, Allerton CMN, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ, Dantonio A, Di L, Eng H, Ferre R, Gajiwala KS, Gibson SA, Greasley SE, Hurst BL, Kadar EP, Kalgutkar AS, Lee JC, Lee J, Liu W, Mason SW, Noell S, Novak JJ, Obach RS, Ogilvie K, Patel NC, Pettersson M, Rai DK, Reese MR, Sammons MF, Sathish JG, Singh RSP, Steppan CM, Stewart AE, Tuttle JB, Updyke L, Verhoest PR, Wei L, Yang Q, Zhu Y (2021) An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 374:1586–1593. https://doi.org/10.1126/science.abl4784
Papadopoulos JS, Agarwala R (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23:1073–1079. https://doi.org/10.1093/bioinformatics/btm076
Pjura PE, Lenhoff AM, Leonard SA, Gittis A (2000) Protein crystallization by design chymotrypsinogen without precipitants. J Mol Biol 300:235–239. https://doi.org/10.1006/jmbi.2000.3851
Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD (2018) The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res 46:D624–D632. https://doi.org/10.1093/nar/gkx1134
Sawa J, Malet H, Krojer T, Canellas F, Ehrmann M, Clausen T (2011) Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope. J Biol Chem 286:30680–30690. https://doi.org/10.1074/jbc.M111.243832
Schechter I, Berger A (1967) On the size of the active site in proteases. I Papain Biochem Biophys Res Commun 27:157–162. https://doi.org/10.1016/s0006-291x(67)80055-x
Shi J, Wei Z, Song J (2004) Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme defining the extra domain as a new target for design of highly specific protease inhibitors. J Biol Chem 279:24765–24773. https://doi.org/10.1074/jbc.M311744200
Shi J, Sivaraman J, Song J (2008) Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J Virol 82:4620–4629. https://doi.org/10.1128/JVI.02680-07
Shi Y, Lei Y, Ye G, Sun L, Fang L, Xiao S, Fu ZF, Yin P, Song Y, Peng G (2018) Identification of two antiviral inhibitors targeting 3C-like serine/3C-like protease of porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus. Vet Microbiol 213:114–122. https://doi.org/10.1016/j.vetmic.2017.11.031
Shindyalov IN, Bourne PE (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng 11:739–747. https://doi.org/10.1093/protein/11.9.739
Smits SL, Snijder EJ, de Groot RJ (2006) Characterization of a torovirus main proteinase. J Virol 80:4157–4167. https://doi.org/10.1128/JVI.80.8.4157-4167.2006
Stroud RM, Kossiakoff AA, Chambers JL (1977) Mechanisms of zymogen activation. Annu Rev Biophys Bioeng 6:177–193. https://doi.org/10.1146/annurev.bb.06.060177.001141
Sun D, Chen S, Cheng A, Wang M (2016) Roles of the picornaviral 3C proteinase in the viral life cycle and host cells. Viruses 8:82. https://doi.org/10.3390/v8030082
Sweeney TR, Roqué-Rosell N, Birtley JR, Leatherbarrow RJ, Curry S (2007) Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J Virol 81:115–124. https://doi.org/10.1128/JVI.01587-06
Tan J, Verschueren KH, Anand K, Shen J, Yang M, Xu Y, Rao Z, Bigalke J, Heisen B, Mesters JR, Chen K, Shen X, Jiang H, Hilgenfeld R (2005) pH-dependent conformational flexibility of the SARS-CoV main proteinase (M(pro)) dimer: molecular dynamics simulations and multiple X-ray structure analyses. J Mol Biol 354:25–40. https://doi.org/10.1016/j.jmb.2005.09.012
Tian X, Lu G, Gao F, Peng H, Feng Y, Ma G, Bartlam M, Tian K, Yan J, Hilgenfeld R, Gao GF (2009) Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV). J Mol Biol 392:977–993. https://doi.org/10.1016/j.jmb.2009.07.062
Tomar S, Johnston ML, St John SE, Osswald HL, Nyalapatla PR, Paul LN, Ghosh AK, Denison MR, Mesecar AD (2015) Ligand-induced dimerization of Middle East respiratory syndrome (MERS) coronavirus nsp5 Protease (3CLpro): implication for nsp5 regulation and the development of antivirals. J Biol Chem 290:19403–19422. https://doi.org/10.1074/jbc.M115.651463
Tsukada H, Blow DM (1985) Structure of alpha-chymotrypsin refined at 1.68 A resolution. J Mol Biol 184:703–711. https://doi.org/10.1016/0022-2836(85)90314-6
Ulferts R, Mettenleiter TC, Ziebuhr J (2011) Characterization of Bafinivirus main protease autoprocessing activities. J Virol 85:1348–1359. https://doi.org/10.1128/JVI.01716-10
Ullrich S, Nitsche C (2020) The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett 30:127377. https://doi.org/10.1016/j.bmcl.2020.127377
Ullrich S, Ekanayake KB, Otting G, Nitsche C (2022) Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir. Bioorg Med Chem Lett 62:128629. https://doi.org/10.1016/j.bmcl.2022.128629
Unoh Y, Uehara S, Nakahara K, Nobori H, Yamatsu Y, Yamamoto S, Maruyama Y, Taoda Y, Kasamatsu K, Suto T, Kouki K, Nakahashi A, Kawashima S, Sanaki T, Toba S, Uemura K, Mizutare T, Ando S, Sasaki M, Orba Y, Sawa H, Sato A, Sato T, Kato T, Tachibana Y (2022) Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J Med Chem 65:6499–6512. https://doi.org/10.1021/acs.jmedchem.2c00117
Uraki R, Kiso M, Iida S, Imai M, Takashita E, Kuroda M, Halfmann PJ, Loeber S, Maemura T, Yamayoshi S, Fujisaki S, Wang Z, Ito M, Ujie M, Iwatsuki-Horimoto K, Furusawa Y, Wright R, Chong Z, Ozono S, Yasuhara A, Ueki H, Sakai-Tagawa Y, Li R, Liu Y, Larson D, Koga M, Tsutsumi T, Adachi E, Saito M, Yamamoto S, Hagihara M, Mitamura K, Sato T, Hojo M, Hattori SI, Maeda K, Valdez R, IASO study team, Okuda M, Murakami J, Duong C, Godbole S, Douek DC, Maeda K, Watanabe S, Gordon A, Ohmagari N, Yotsuyanagi H, Diamond MS, Hasegawa H, Mitsuya H, Suzuki T, Kawaoka Y (2022) Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature. 607:119–127. https://doi.org/10.1038/s41586-022-04856-1
van Aken D, Snijder EJ, Gorbalenya AE (2006) Mutagenesis analysis of the nsp4 main proteinase reveals determinants of arterivirus replicase polyprotein autoprocessing. J Virol 80:3428–3437. https://doi.org/10.1128/JVI.80.7.3428-3437.2006
van Tilbeurgh H, Egloff MP, Martinez C, Rugani N, Verger R, Cambillau C (1993) Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography. Nature 362:814–820. https://doi.org/10.1038/362814a0
Wang YC, Yang WH, Yang CS, Hou MH, Tsai CL, Chou YZ, Hung MC, Chen Y (2020) Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug. Am J Cancer Res 10:2535–2545
Wang Z, Sun H, Shen C, Hu X, Gao J, Li D, Cao D, Hou T (2020) Combined strategies in structure-based virtual screening. Phys Chem Chem Phys 22:3149–3159. https://doi.org/10.1039/c9cp06303j
Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T (2004) Crystal structure of the DegS stress sensor. How a PDZ domain recognizes misfolded protein and activates a protease. Cell 117:483–494. https://doi.org/10.1016/s0092-8674(04)00454-4
Woo PC, Huang Y, Lau SK, Yuen KY (2010) Coronavirus genomics and bioinformatics analysis. Viruses 2:1804–1820. https://doi.org/10.3390/v2081803
Wu CG, Cheng SC, Chen SC, Li JY, Fang YH, Chen YH, Chou CY (2013) Mechanism for controlling the monomer-dimer conversion of SARS coronavirus main protease. Acta Crystallogr D Biol Crystallogr 69:747–755. https://doi.org/10.1107/S0907444913001315
Xia B, Kang X (2011) Activation and maturation of SARS-CoV main protease. Protein Cell 2:282–290. https://doi.org/10.1007/s13238-011-1034-1
Xu S, Zhou J, Chen Y, Tong X, Wang Z, Guo J, Chen J, Fang L, Wang D, Xiao S (2020) Characterization of self-processing activities and substrate specificities of porcine torovirus 3C-like protease. J Virol 94:e01282-e1320. https://doi.org/10.1128/JVI.01282-20
Xue X, Yang H, Shen W, Zhao Q, Li J, Yang K, Chen C, Jin Y, Bartlam M, Rao Z (2007) Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J Mol Biol 366:965–975. https://doi.org/10.1016/j.jmb.2006.11.073
Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, Li J, Zhou Z, Ding Y, Zhao Q, Zhang XC, Liao M, Bartlam M, Rao Z (2008) Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J Virol 82:2515–2527. https://doi.org/10.1128/JVI.02114-07
Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z (2003) The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA 100:13190–13195. https://doi.org/10.1073/pnas.1835675100
Ye G, Deng F, Shen Z, Luo R, Zhao L, Xiao S, Fu ZF, Peng G (2016) Structural basis for the dimerization and substrate recognition specificity of porcine epidemic diarrhea virus 3C-like protease. Virology 494:225–235. https://doi.org/10.1016/j.virol.2016.04.018
Yi J, Peng J, Yang W, Zhu G, Ren J, Li D, Zheng H (2021) Picornavirus 3C - a protease ensuring virus replication and subverting host responses. J Cell Sci 134:jcs253237. https://doi.org/10.1242/jcs.253237
Ziebuhr J, Heusipp G, Siddell SG (1997) Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J Virol 71:3992–3997. https://doi.org/10.1128/JVI.71.5.3992-3997.1997
Ziebuhr J, Bayer S, Cowley JA, Gorbalenya AE (2003) The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J Virol 77:1415–1426. https://doi.org/10.1128/jvi.77.2.1415-1426.2003
Funding
This research was supported by the Research Support Project for Life Science and Drug Discovery (project ID: JP22ama121023) from the Japan Agency for Medical Research and Development (AMED).
Author information
Authors and Affiliations
Contributions
The computational analyses were performed by all authors. The first draft was written by AK, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kidera, A., Moritsugu, K., Ekimoto, T. et al. Functional dynamics of SARS-CoV-2 3C-like protease as a member of clan PA. Biophys Rev 14, 1473–1485 (2022). https://doi.org/10.1007/s12551-022-01020-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-022-01020-x