Abstract
Autism spectrum disorder (ASD) is a neurological and developmental disorder and its early diagnosis is a challenging task. The dynamic brain network (DBN) offers a wealth of information for the diagnosis and treatment of ASD. Mining the spatio-temporal characteristics of DBN is critical for finding dynamic communication across brain regions and, ultimately, identifying the ASD diagnostic biomarker. We proposed the dgEmbed-KNN and the Aggregation-SVM diagnostic models, which use the spatio-temporal information from DBN and interactive information among brain regions represented by dynamic graph embedding. The classification accuracies show that dgEmbed-KNN model performs slightly better than traditional machine learning and deep learning methods, while the Aggregation-SVM model has a very good capacity to diagnose ASD using aggregation brain network connections as features. We discovered over- and under-connections in ASD at the level of dynamic connections, involving brain regions of the postcentral gyrus, the insula, the cerebellum, the caudate nucleus, and the temporal pole. We also found abnormal dynamic interactions associated with ASD within/between the functional subnetworks, including default mode network, visual network, auditory network and saliency network. These can provide potential DBN biomarkers for ASD identification.
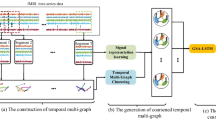
Graphical Abstract







Similar content being viewed by others
Data availability
The data that support the findings of this study are openly available in ABIDE online repository (ref. [24]).
References
Bahathiq RA, Banjar H, Bamaga AK et al (2022) Machine learning for autism spectrum disorder diagnosis using structural magnetic resonance imaging: promising but challenging. Front Neuroinform 16:949926. https://doi.org/10.3389/fninf.2022.949926
Kaur P, Kaur A (2023) Review of progress in diagnostic studies of autism spectrum disorder using neuroimaging. Interdiscip Sci 15(1):111–130. https://doi.org/10.1007/s12539-022-00548-6
Maenner MJ, Warren Z, Williams AR et al (2023) Prevalence and characteristics of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2020. MMWR Surveill Summ 72(2):1–14
Feng W, Liu G, Zeng K et al (2022) A review of methods for classification and recognition of ASD using fMRI data. J Neurosci Methods 368:109456. https://doi.org/10.1016/j.jneumeth.2021.109456
Reiter MA, Jahedi A, Jac Fredo AR et al (2021) Performance of machine learning classification models of autism using resting-state fMRI is contingent on sample heterogeneity. Neural Comput Appl 33(8):3299–3310. https://doi.org/10.1007/s00521-020-05193-y
Spera G, Retico A, Bosco P et al (2019) Evaluation of altered functional connections in male children with autism spectrum disorders on multiple-site data optimized with machine learning. Front Psychiatry 10:620. https://doi.org/10.3389/fpsyt.2019.00620
Wang CH, Xiao ZY, Wang BY et al (2019) Identification of autism based on SVM-RFE and stacked sparse auto-encoder. IEEE Access 7:118030–118036. https://doi.org/10.1109/access.2019.2936639
Grana M, Silva M (2021) Impact of machine learning pipeline choices in autism prediction from functional connectivity data. Int J Neural Syst 31(4):2150009. https://doi.org/10.1142/S012906572150009X
Shi C, Xin X, Zhang J (2021) Domain adaptation using a three-way decision improves the identification of autism patients from multisite fMRI data. Brain Sci. https://doi.org/10.3390/brainsci11050603
Chaitra N, Vijaya PA, Deshpande G (2020) Diagnostic prediction of autism spectrum disorder using complex network measures in a machine learning framework. Biomed Signal Process Control 62:102099. https://doi.org/10.1016/j.bspc.2020.102099
Mostafa S, Tang L, Wu F-X (2019) Diagnosis of autism spectrum disorder based on eigenvalues of brain networks. IEEE Access 7:128474–128486. https://doi.org/10.1109/access.2019.2940198
Kazeminejad A, Sotero RC (2020) The importance of anti-correlations in graph theory based classification of autism spectrum disorder. Front Neurosci 14:676. https://doi.org/10.3389/fnins.2020.00676
Carboni L, Achard S, Dojat M (2021) Network embedding for brain connectivity. In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), pp 1722–1725
Wang ML, Lian CF, Yao DR et al (2020) Spatial-temporal dependency modeling and network hub detection for functional MRI analysis via convolutional-recurrent network. IEEE Trans Biomed Eng 67(8):2241–2252. https://doi.org/10.1109/TBME.2019.2957921
Zhao F, Chen ZY, Rekik I et al (2020) Diagnosis of autism spectrum disorder using central-moment features from low- and high-order dynamic resting-state gunctional connectivity networks. Front Neurosci 14:258. https://doi.org/10.3389/fnins.2020.00258
Liu J, Sheng Y, Lan W et al (2020) Improved ASD classification using dynamic functional connectivity and multi-task feature selection. Pattern Recog Lett 138:82–87. https://doi.org/10.1016/j.patrec.2020.07.005
Yan WZ, Zhang H, Sui J et al (2018) Deep chronnectome learning via full bidirectional long short-term memory networks for MCI diagnosis. Med Image Comput Comput Assist Interv 11072:249–257. https://doi.org/10.48550/arXiv.1808.10383
Ji J, Chen Z, Yang C (2022) Convolutional neural network with sparse strategies to classify dynamic functional connectivity. IEEE J Biomed Health Inform 26(3):1219–1228. https://doi.org/10.1109/JBHI.2021.3100559
Li XX, Zhou Y, Dvornek N et al (2021) BrainGNN: interpretable brain graph beural betwork for fMRI analysis. Med Image Anal 74:102233. https://doi.org/10.1016/j.media.2021.102233
Jiang H, Cao P, Xu MY et al (2020) Hi-GCN: a hierarchical graph convolution network for graph embedding learning of brain network and brain disorders prediction. Comput Biol Med 127:104096. https://doi.org/10.1016/j.compbiomed.2020.104096
Cao P, Wen G, Liu X et al (2022) Modeling the dynamic brain network representation for autism spectrum disorder diagnosis. Med Biol Eng Comput 60(7):1897–1913. https://doi.org/10.1007/s11517-022-02558-4
Xing XD, Li QF, Yuan MY et al (2021) DS-GCNs: connectome classification using dynamic spectral graph convolution networks with assistant task training. Cereb Cortex 31(2):1259–1269. https://doi.org/10.1093/cercor/bhaa292
Xue G, Zhong M, Li J et al (2022) Dynamic network embedding survey. Neurocomputing 472:212–223. https://doi.org/10.1016/j.neucom.2021.03.138
Di Martino A, Yan CG, Li Q et al (2014) The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 19(6):659–667. https://doi.org/10.1038/mp.2013.78
Abraham A, Milham MP, Di Martino A et al (2017) Deriving reproducible biomarkers from multi-site resting-state data: an autism-based example. Neuroimage 147:736–745. https://doi.org/10.1016/j.neuroimage.2016.10.045
Craddock C, Benhajali Y, Chu C et al (2013) The neuro bureau preprocessing initiative: open sharing of preprocessed neuroimaging data and derivatives. Front Neuroinform. https://doi.org/10.3389/conf.fninf.2013.09.00041
Craddock RC, James GA, Holtzheimer PE 3rd et al (2012) A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp 33(8):1914–1928. https://doi.org/10.1002/hbm.21333
Preti MG, Bolton TA, Van De Ville D (2017) The dynamic functional connectome: state-of-the-art and perspectives. Neuroimage 160:41–54. https://doi.org/10.1016/j.neuroimage.2016.12.061
Beladev M, Rokach L, Katz G et al (2020) tdGraphEmbed: temporal dynamic graph-level embedding. In: Proceedings of the 29th ACM International Conference on Information & Knowledge Management
Grover A, Leskovec J (2016) node2vec: scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp 855–864
Le Q, Mikolov T (2014) Distributed representations of sentences and documents. Int Conf Mach Learn 32:II-1188–II−1196. https://doi.org/10.48550/arXiv.1405.4053
Bunke H, Shearer K (1998) A graph distance metric based on the maximal common subgraph. Pattern Recog Lett 19(3–4):255–259. https://doi.org/10.1016/S0167-8655(97)00179-7
Pan X, Zuallaert J, Wang X et al (2021) ToxDL: deep learning using primary structure and domain embeddings for assessing protein toxicity. Bioinformatics 36(21):5159–5168. https://doi.org/10.1093/bioinformatics/btaa656
Zhang T, Ramakrishnan R, Livny M (1996) BIRCH: an efficient data clustering method for very large databases. Sigmod Rec 25(2):103–114. https://doi.org/10.1145/233269.233324
Joe H, Ward J (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58(301):236–244. https://doi.org/10.1080/01621459.1963.10500845
Eslami T, Mirjalili V, Fong A et al (2019) ASD-DiagNet: a hybrid learning approach for detection of autism spectrum disorder using fMRI data. Front Neuroinform 13:70. https://doi.org/10.3389/fninf.2019.00070
Wen G, Cao P, Bao H et al (2022) MVS-GCN: a prior brain structure learning-guided multi-view graph convolution network for autism spectrum disorder diagnosis. Comput Biol Med 142:105239. https://doi.org/10.1016/j.compbiomed.2022.105239
Xia M, Wang J, He Y (2013) BrainNet viewer: a network visualization tool for human brain connectomics. PLoS ONE 8(7):e68910. https://doi.org/10.1371/journal.pone.0068910
Tzourio-Mazoyer N, Landeau B, Papathanassiou D et al (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15(1):273–289. https://doi.org/10.1006/nimg.2001.0978
Huang YT, Zhang BL, Cao J et al (2020) Potential locations for noninvasive brain stimulation in treating autism spectrum disorders—a functional connectivity study. Front Psychiatry 11:388. https://doi.org/10.3389/fpsyt.2020.00388
Ramos TC, Balardin JB, Sato JR et al (2019) Abnormal cortico-cerebellar functional connectivity in autism spectrum disorder. Front Syst Neurosci 12:74. https://doi.org/10.3389/fnsys.2018.00074
Mizuno Y, Kagitani-Shimono K, Jung M et al (2019) Structural brain abnormalities in children and adolescents with comorbid autism spectrum disorder and attention-deficit/hyperactivity disorder. Transl Psychiatry 9(1):332. https://doi.org/10.1038/s41398-019-0679-z
Ortug A, Guo Y, Feldman HA et al (2022) Human fetal brain magnetic resonance imaging (MRI) tells future emergence of autism spectrum disorders. FASEB J. https://doi.org/10.1096/fasebj.2022.36.S1.R2353
Yamada T, Itahashi T, Nakamura M et al (2016) Altered functional organization within the insular cortex in adult males with high-functioning autism spectrum disorder: evidence from connectivity-based parcellation. Mol Autism 7:41. https://doi.org/10.1186/s13229-016-0106-8
Xu JP, Wang HW, Zhang L et al (2018) Both hypo-connectivity and hyper-connectivity of the insular subregions associated with severity in children with autism spectrum disorders. Front Neurosci 12:234. https://doi.org/10.3389/fnins.2018.00234
Butera C, Kaplan J, Kilroy E et al (2023) The relationship between alexithymia, interoception, and neural functional connectivity during facial expression processing in autism spectrum disorder. Neuropsychologia 180:108469. https://doi.org/10.1016/j.neuropsychologia.2023.108469
Yerys BE, Gordon EM, Abrams DN et al (2015) Default mode network segregation and social deficits in autism spectrum disorder: evidence from non-medicated children. NeuroImage Clin 9:223–232. https://doi.org/10.1016/j.nicl.2015.07.018
Al-Hiyali MI, Yahya N, Faye I et al (2021) Identification of autism subtypes based on wavelet coherence of BOLD fMRI signals using convolutional neural network. Sensors. https://doi.org/10.3390/s21165256
Qiu T, Chang C, Li Y et al (2016) Two years changes in the development of caudate nucleus are involved in restricted repetitive behaviors in 2–5-year-old children with autism spectrum disorder. Dev Cogn Neurosci 19:137–143. https://doi.org/10.1016/j.dcn.2016.02.010
Long Z, Duan X, Mantini D et al (2016) Alteration of functional connectivity in autism spectrum disorder: effect of age and anatomical distance. Sci Rep 6:26527. https://doi.org/10.1038/srep26527
Bednarz HM, Kana RK (2019) Patterns of cerebellar connectivity with intrinsic connectivity networks in autism spectrum disorders. J Autism Dev Disord 49(11):4498–4514. https://doi.org/10.1007/s10803-019-04168-w
Xie J, Wang L, Webster P et al (2022) Identifying visual attention reatures accurately discerning between autism and typically developing: a deep learning framework. Interdiscip Sci 14(3):639–651. https://doi.org/10.1007/s12539-022-00510-6
Menon V (2011) Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn Sci 15(10):483–506. https://doi.org/10.1016/j.tics.2011.08.003
Acknowledgements
We thank Professor Li Liu from the Big Data Center of Affiliated Hospital of Jiangnan University for her support and assistance in data collection, preprocessing, and analysis of potential DBN biomarkers identified in this study. This work is supported in part by the National Natural Science Foundation of China under Grant 62161050, and in part by the Postgraduate Research & Practice Innovation Program of Jiangsu Province under Grant KYCX22_2419.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Y., Wang, H. & Ding, Y. The Dynamical Biomarkers in Functional Connectivity of Autism Spectrum Disorder Based on Dynamic Graph Embedding. Interdiscip Sci Comput Life Sci 16, 141–159 (2024). https://doi.org/10.1007/s12539-023-00592-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-023-00592-w