Abstract
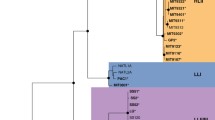
Prochlorococcus is the smallest known oxygenic phototrophic marine cyanobacterium dominating the mid-latitude oceans. Physiologically and genetically distinct P. marinus isolates from many oceans in the world were assigned two different groups, a tightly clustered high-light (HL)-adapted and a divergent low-light (LL-) adapted clade. Phylogenetic analysis of this cyanobacterium on the basis of 16S rRNA and other conserved genes did not show consistency with its phenotypic behavior. We analyzed phylogeny of this genus on the basis of complete genome sequences through genome alignment, overlapping-gene content and gene-order approach. Phylogenetic tree of P. marinus obtained by comparing whole genome sequences in contrast to that based on 16S rRNA gene, corresponded well with the HL/LL ecotypic distinction of twelve strains and showed consistency with phenotypic classification of P. marinus. Evidence for the horizontal descent and acquisition of genes within and across the genus was observed. Many genes involved in metabolic functions were found to be conserved across these genomes and many were continuously gained by different strains as per their needs during the course of their evolution. Consistency in the physiological and genetic phylogeny based on whole genome sequence is established. These observations improve our understanding about the adaptation and diversification of these organisms under evolutionary pressure.
Similar content being viewed by others
References
Angiuoli, S.V., Hotopp, J.C.D., Salzberg, S.L., Tettelin, H. 2011. Improving pan-genome annotation using whole genome multiple alignment. BMC Bioinformatics 12, 272.
Auch, A.F., Klenk, H., Göker, M. 2010. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2, 142–148.
Bryant. 2003. The beauty in small things revealed. Proc Natl Acad Sci USA 100, 9647–9649.
Bryant, D., Moulton, V. 2004. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21, 255–256.
Chevenet, F., Brun, C., Banuls, A.L., Jacq, B., Chisten, R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439.
Darling, A.C., Mau, B., Blattner, F.R., Perna, N.T. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14(7), 1394–1403.
Darling, A.E., Mau, B., Perna, N.T. 2010. progressive-Mauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 5(6), e11147.
Darling, A.E., Miklo’s, I., Ragan, M. A. 2008. Dynamics of Genome Rearrangement in Bacterial Populations. PLoS Genet 4(7), e1000128.
Darling, A.E., Treangen, T.J., Messeguer, X., Perna, N.T. 2007. Analyzing patterns of microbial evolution using the mauve genome alignment system. Methods Mol Biol 396, 135–152.
Delcher, A.L., Phillippy, A., Carlton, J., Salzberg, S.L. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30, 2478–2483.
Dikow, R.B. 2011. Genome-level homology and phylogeny of Shewanella (Gammaproteobacteria: lteromonadales: Shewanellaceae). BMC Genomics 12, 237.
Dufresne, A., Garczarek, L., Partensky, F. 2005. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol 6, R14.
Dufresne, A., Salanoubat, M., Partensky, F., Artiguenave, F., Axmann, I.M., Barbe, V., Duprat, S., Galperin, M.Y., Koonin, E.V., Le Gall, F., Makarova, K.S., Ostrowski, M., Oztas, S., Robert, C., Rogozin, I.B., Scanlan, D.J., Tandeau de Marsac, N., Weissenbach, J., Wincker, P., Wolf, Y.I., Hess, W.R. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Natl Acad Sci USA 100, 10020–10025.
Edgar, R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32, 1792–1797.
Eisen, J.A. 2000. Assessing evolutionary relationships among microbes from whole-genome analysis. Curr Opin Microbiol 3, 475–480.
Henz, S.R., Huson, D.H., Auch, A.F., Nieselt-Struwe, K., Schuster, S.C. 2005. Whole-genome prokaryotic phylogeny. Bioinformatics 21, 2329–2335.
Hess, W.R., Rocap, G., Ting, C.S., Larimer, F., Stilwagen, S., Lamerdin, J., Chisholm, S.W. 2001. The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosynth Res 70, 53–71.
Huson, D.H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14, 68–73.
Jiang, L., Lin, K., Lu, C.L. 2008. OGtree: a tool for creating genome trees of prokaryotes based on overlapping genes. Nucl Acids Res 36, W475–W480.
Kettler, C.G., Martiny, A.C., Huang, K., Zucker, J., Coleman, M.L., Rodrigue, S., Chen, F., Lapidus, A., Ferriera, S.., Johnson, J., Steglich, C., Church, G.M., Richardson, P., Chisholm, S.W. 2007. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3, e231.
Laing, C., Buchanan, C., Taboada, E.N., Zhang, Y., Kropinski, A., Villegas, A., Thomas, J.E., Gannon, V.P.J. 2010. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11, 461.
Lawrence, J.G., Ochman, H. 1997. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol 44, 383–397.
Lawrence, J.G., Ochman, H. 1998. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA 95, 9413–9417.
Luo, H., Shi, J., Arndt, W., Tang, J., Friedman, R. 2008. Gene Order phylogeny of the genus Prochlorococcus. PLoS ONE 3, e3837.
Luo, Y., Fu, C., Zhang, D.Y., Lin, K. 2006. Overlapping genes as rare genomic markers: the phylogeny of g-Proteobacteria as a case study. Trends Genet 22, 593–596.
Markowitz, V.M., Chen, A.I., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., Ratner, A., Jacob, B., Huang, J., Williams, P., Huntemann, M., Anderson, I., Mavromatis, K., Ivanova, N.N., Kyrpides, N.C. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucl Acids Res. 40, D115–D122.
Martiny, A.C., Coleman, M.L., Chisholm, S.W. 2006. Phosphate acquisition genes in Prochlorococcus ecoytypes: evidence for genome wide adaptation. Proc Natl Acad Sci USA 103, 12552–12557.
Miele, V., Penel, S., Duret, L. 2011. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12, 116.
Moore, L.R., Goericke, R.E., Chisholm, S.W. 2002. Utilization of different nitrogen sources by the marine cyanobacterial, Prochlorococcus and Synechococcus. Limnol Oceanogr 47, 989–996.
Moore, L.R., Rocap, G., Chisholm, S.W. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467.
Nakamura, Y., Itoh, T., Matsuda, H., Gojobor, T. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet 36, 760–766.
Partensky, F., Garczarek, L. 2010. Prochlorococcus: advantages and limits of minimalism. Ann Rev Mar Sci 2, 305–331.
Partensky, F., Hess, W.R., Vaulot, D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63, 106–127.
Paul, S., Dutta, A., Bag, S.K., Das, S. 2010. Distinct, ecotype-specific genome and proteome signatures in the marine cyanobacteria Prochlorococcus. BMC Genomics 11, 103.
Qi, J., Wang, B., Hao, B.I. 2004. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J Mol Evol 58, 1–11.
Rocap, G., Distel, D.L., Waterbury, J.B., Chisholm, S.W. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S–23S ribosomal DNA internal transcribed spacer sequences. Appl Env Microbiol 68, 1180–1191.
Rocap, G., Larimer, F.W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N.A., Arellano, A., Coleman, M., Hauser, L., Hess, W.R., Johnson, Z.I., Land, M., Lindell, D., Post, A.F., Regala, W., Shah, M., Shaw, S.L., Steglich, C., Sullivan, M.B., Ting, C.S., Tolonen, A., Webb, E.A., Zinser, E.R., Chisholm, S.W. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047.
Sims, G.E., Jun, S., Wu, G.A., Kim, S. 2009. Wholegenome phylogeny of mammals: evolutionary information in genic and nongenic regions. Proc Natl Acad Sci USA 106, 17077–17082.
Snel, B., Bork, P., Huynen, M.A. 1999. Genome phylogeny based on gene content. Nat Genet 21, 108–110.
Snel, B., Huynen, M.A., Dutilh, B.E. 2005. Genome trees and the nature of genome evolution. Ann Rev Microbiol 59, 191–209.
Sullivan, M.B., Coleman, M.L., Weigele, P., Rohwer, F., Chisholm, S.W. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3, e144.
Sullivan, M.B., Waterbury, J.B., Chisholm, S.W. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424, 1047–1051.
Sun, Z. 2011. New genomic approaches reveal the process of genome reduction in Prochlorococcus. Open Access Dissertations. Paper 352.
Suyama, M., Bork, P. 2001. Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends Genet 17, 10–13.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739.
Tettelin, H., Riley, D., Cattuto, C., Medini, D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 12, 472–477.
Vishnoi, A., Roy, R., Prasad, H.K., Bhattacharya, A. 2010. Anchor-Based Whole Genome Phylogeny (ABWGP): a tool for inferring evolutionary relationship among closely related microorganisms. PLoS ONE 5, e14159.
Wolf, Y.I., Rogozin, I.B., Grishin, N.V., Tatusov, R.L., Koonin, E.V. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol Biol. 1, 8.
Zeidner, G., Bielawski, J.P., Shmoish, M., Scanlan, D.J., Sabehi, G., Beja, O. 2005. Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ Microbiol 7, 1505–1513.
Zhaxybayeva, O., Doolittle, W.F., Papke, R.T., Gogarten, J.P. 2009. Intertwined evolutionary histories of marine Synechococcus and Prochlorococcus marinus. Genome Biol Evol 1, 325–339.
Zhaxybayeva, O., Gogarten, J.P., Charlebois, R.L., Doolittle, W.F., Papke, R.T. 2006. Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res 16, 1099–1108.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prabha, R., Singh, D.P., Gupta, S.K. et al. Whole genome phylogeny of Prochlorococcus marinus group of cyanobacteria: genome alignment and overlapping gene approach. Interdiscip Sci Comput Life Sci 6, 149–157 (2014). https://doi.org/10.1007/s12539-013-0024-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-013-0024-9