Abstract
Background
Aflatoxin B1 (AFB1) a mycotoxin produced by Aspergillus flavus and A. parasiticus is a potent carcinogen and causative agent of hepatocellular carcinoma (HCC). It is a food contaminant which presents a major risk to human health. AFB1 contamination poses a significant economic burden, as 25% of the world's food crops need to be destroyed annually. The mechanism of action (MOA) of aflatoxins remains to be fully elucidated. Recent findings suggest that AFB1 mediated endocrine disruption may occur in the population of regions with high contamination, even without evidence of direct dietary intake.
Objective
An integrative systems biology approach was undertaken to decipher the estrogenic component of the mechanism of action (MOA) of AFB1.
Methods
Molecular Docking and Molecular dynamics simulations were performed to examine the binding affinity of AFB1 and its metabolite aflatoxin Q1 (AFQ1) with the Estrogen Receptors (ERs). Differential gene expression (DGE), gene ontology (GO) and pathway analyses were carried out on hepatic transcriptomic data generated from in vivo AFB1 exposures. In parallel exposures to the synthetic estrogen ethinylestradiol (EE2) were examined for overlapping effects. Finally, protein–protein interaction (PPI) network analysis assessed the involvement of estrogen responsive targets (ERTs) associated with aflatoxin exposure.
Results
The free energies of binding affinity and estimated equilibrium dissociation constants (KD) demonstrated that AFB1 and AFQ1 can interact with the ERα and ERβ. DGE and GO analyses highlighted overlap in the responses between AFB1 and EE2 treatments with the activation of key processes involved in estrogenic signaling. PPI network analyses after AFBI exposure revealed a dynamic response to AFB1 treatments with the solid involvement of ERTs in regulatory networks.
Conclusions
This study revealed molecular interactions between aflatoxins (AFB1, AFQ1) and ERs in addition to overlap in differentially expressed genes and biological processes following AFB1 and EE2 exposures. The estrogenic components at the core of the PPI networks suggest that ER-mediated signaling pathways are a major component in the MOA of aflatoxins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aflatoxins are a family of secondary metabolites or mycotoxins produced by different species belonging to the Aspergillus genus. Aspergillus flavus and A. parasiticus are the best studied species that produce Aflatoxins. Although A. parasiticus is better adapted to grow in soils, its distribution is more limited than A. flavus and it mainly produces aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), aflatoxin G1 (AFG1) and aflatoxin G2 (AFG2). A. flavus has a broader host range and contaminates the aerial parts of plants producing AFB1 and AFB2 leading to more critical food contamination (EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020). Aspergillus growth and relative aflatoxin contamination in crops, foods and animal feed can occur at any time pre- and post-harvest stage, making it difficult to control the contamination rate of foods (Food Safety Digest, Department of Food safety and Zoonoses). Moreover, high humidity and temperatures enhance fungal growth posing challenges in nations with warmer climates and particularly developing countries, to safely store harvested foods. Consequently, people living in these environments are more prone to aflatoxin contaminations and acute intoxications (aflatoxicosis) cases (EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020; Rushing and Selim 2019).
In recent years aflatoxin outbreaks have been recorded in South-European regions including Italy in 2003 and Serbia in 2012 (Battilani et al. 2016; EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020). These events suggest that climate change induced increases in temperature and humidity could expand Aspergillus habitats and high-risk contamination temperate areas (e.g., Europe). Two predictive models based on + 2 and + 5 °C climate changes scenarios respectively have projected an increase in maize and wheat contamination risk in numerous European nations (Battilani et al. 2016). Additionally, evidence suggests that the combination of climate change factors such as elevated temperature, CO2 levels in the air and water shortage can cause a switch from Fusarium and fumonisins to A. flavus and aflatoxin contamination (Kolawole et al. 2021; Medina et al. 2014) and significantly increase expression of the key regulators of Aflatoxin B1 production aflD and aflR (Kolawole et al. 2021; Medina et al. 2014).
AFB1 is classified as a group 1 carcinogen for humans by the International Agency for Research on Cancer (Centre international de recherche sur le cancer 2002). Although the Codex Alimentarius has issued specific guidelines for maximum permitted levels of total aflatoxins in food (1 μg/kg for cereal-based baby food to 20 μg/kg for cereals used for further processing) (CCCF14 2020), the total aflatoxin limits differ in many parts of the world. For example, up to 20 μg/kg of total aflatoxins in food are allowed in the United States (Eskola et al. 2020; FDA 2021) and up to 15 μg / kg as an industry standard in peanut production (US Government 2002). The European Union regulates the levels of aflatoxin contamination by distinguishing between total aflatoxin content and AFB1, allowing up to 8 μg/kg of AFB1 and 15 μg/kg of total aflatoxin contamination in further processed food and up to 2 μg/kg of AFB1 and 4 μg/kg of total aflatoxins in ready to eat peanuts (Eskola et al. 2020; European Commission 2006a, 2006b).
AFB1 exposure is a causative agent of Hepatocellular Carcinoma (HCC) (EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020; Rushing and Selim 2019). Aflatoxin B1 exerts toxic effects, primarily on the liver. The primary carcinogenic and genotoxic effects act through two distinct mechanisms (Benkerroum 2020; Moore et al. 2018). The first and best known are the effects of the highly reactive metabolites AFB1-exo and -endo 8,9 epoxide (Epo-AFB1) produced by CYP450 enzymes. The second genotoxic effect acts via an oxidative stress response. AFB1 metabolism in the liver by CYP450 releases excess amounts of reactive oxygen species (ROS). (Benkerroum 2020; Moore et al. 2018).
Considering the similarities between the chemical structures of AFB1 and estradiol, the binding affinity between AFB1 and bovine ERs has been previously examined using in vitro binding assays (Blankenship et al. 1982; Kyrein 1974). Although both studies observed no affinity between AFB1 and the receptors, Kyrein (1974) observed interactions between elevated concentrations of the metabolite aflatoxin M1 (AFM1) and ERs (Kyrein 1974). The study by Blankenship et al. (1982) found that the AFB1 metabolite Aflatoxicol (AFL) bound the ERs (Blankenship et al. 1982). Deregulation of gene expression by AFB1 and AFM1 led to perturbation of biological pathways inherent to steroid hormone synthesis and estrogen signaling suggesting an inherent estrogenic component in its mechanism of action (Marchese et al. 2018). AFB1 and its metabolite AFL, can cross the placenta and be reconverted to AFB1 (EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020; Huuskonen et al. 2013; Storvik et al. 2011). Furthermore after a single dose of AFB1 in gestating mice, the same DNA adducts (with N7 of Guanine and AFB1-formamidopyrimidine) were found in both mother and fetus (EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020). The effects of AFB1 exposure in placental tissue caused impairment of hormone synthesis and homeostasis, suggesting that AFB1 is a potential endocrine disruptor (EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020; Huuskonen et al. 2013; Storvik et al. 2011). Additional evidence indicating the potentially endocrine-disrupting effects of AFB1 on reproductive tissue derived from mice testis exposed to AFB1 showed the deregulation of expression of Estrogen Receptor α (ERα) (Zamir-Nasta et al. 2017).
Studies in AFB1-exposed goats revealed altered levels of estradiol and progesterone in circulating blood, suggesting effective hormonal disruption by the mycotoxin (Kourousekos et al. 2018; Kourousekos and Theodosiadou 2015). Additionally, the selective estrogen receptor modulator Tamoxifen prevented AFB1 from interacting with enzymes involved in placenta hormone production (CYP19A1) and decreased accumulation of its metabolite Aflatoxicol (Storvik et al. 2011). Finally, a recent study conducted on flour mill workers in Egypt uncovered disruption of several hormones among those who work in environments contaminated by A. flavus (Beshir et al. 2020). This suggests that AFB1-mediate endocrine disruption may occur in highly contaminated locations in the absence of direct dietary intake.
We hypothesized that Aflatoxin B1 and/or its metabolites can directly interact with the nuclear and membrane estrogen receptors (ERs) α and β, dysregulating gene expression and perturbing biological pathways related to this class of receptors. To test this hypothesis and assess the endocrine disrupting effects of AFB1 and its metabolites we applied an integrative system biology approach that considered structural information from investigating ERs and aflatoxin binding, along with transcriptomic data and protein–protein interaction (PPI) network analysis.
Methods
Ligands and Receptors Molecular Structures
Ligand selection was guided by hepatic metabolic pathways (EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020; Marchese et al. 2018; Rushing and Selim 2019; Storvik et al. 2011). A total of 5 samples, 2 positive controls and 1 negative control were selected (Fig. 1). The samples were AFB1, Aflatoxin M1 (AFM1), Aflatoxin Q1 (AFQ1), Aflatoxicol (AFL) and Aflatoxin B1-epoxide (Epo-AFB1). EE2 and the mycoestrogen Zearalenone (ZEA) were included as positive estrogenic controls and Rifampin (RIF) as a negative estrogenic control.
Molecular structure of the compounds used for Molecular Docking simulations. The presence of an aromatic ring is observed in every compound. Apart from ZEA and RIF all compounds are composed of four to five linked rings, features that increase rigidity of the structures. The presence of carbons in the sp2 hybridization, in addition to the aromatic ring and the closed conformation suggests a possible planar conformation of AFB1 and its metabolites. The molecular weights of mycotoxins range from 312.3 to 328.3 g/mol. The molecular weight of EE2 is 296.4 g/mol. RIF, the negative control has a molecular weight of 822.9 g/mol. Structure images were obtained with MolView (MolView.Org)
From the Protein Data Bank (PDB) the molecular structures for human estrogen receptor α (ERα) (Brzozowski et al. 1997), ID:1ERE, and human estrogen receptor β (ERβ) (Mewshaw et al. 2007), ID:2NV7, were downloaded as templates for docking simulations. Receptor preparation, consisted of the selection of chain A, hydrogen addition and conversion to PDBQT file format. This was performed using UCSF Chimera (version 1.14) (Pettersen et al. 2004) and AutoDockTools (ADT) (version 1.5.6) (Morris et al. 2009).
Molecular Docking Simulations
Molecular docking simulation were performed with AutoDock4 (version 4.2.6) (Morris et al. 2009). Every previously selected ligand was docked to both the estrogen receptors structures in a cubical grid box centered on the original ligand co-crystallized with the protein and a side of 22.5 Å, 60 points × 0.375 Å spacing. AutoDock4 was run using the Lamarckian Genetic Algorithm (LGA) for 200 trials with a maximum of 1,000,000 energy evaluations. All the routable bonds of each ligand were set as flexible, the residues of the binding pocket of each receptor were set as rigid.
Molecular Dynamics Simulations
To better evaluate the affinity and stability of the ligands, the docked complexes of both the receptors with AFB1, AFQ1 (optimal ligand) and EE2 (positive control), a total of six complexes, were subjected to molecular dynamics simulation after the complete rebuilding of the full dimeric structure. The precise protonation at a pH of 7.2 (Casey et al. 2010) was carried out by submitting each complex to ProteinPrepare (https://playmolecule.com/proteinPrepare/) (Martínez-Rosell et al. 2017). All the simulations were carried out using GROMACS (version 2021.4)(Abraham et al. 2015; Páll et al. 2015) and the improved side-chain torsion AMBER force field (ff99SB-ILDN) (Lindorff-Larsen et al. 2010) to describe protein parameters. The production phase consisted of 50 ns of NPT simulation set at atmospheric pressure and 300 K degrees for each complex. Periodic boundary conditions (PBCs) were applied in all directions using a neighbour searching grid type and setting at 1.0 nm the cut-off distance for the short-range neighbour list. The temperature was kept at 300 K using the V-rescale thermostat algorithm (Lee et al. 2014) and the Parrinello-Rahman barostat was used to maintain pressure at 1 bar (Parrinello and Rahman 1981). During these steps, protein and ligand as well as water and ions were coupled to their own temperature and pressure while a full positional constrains was imposed on the heavy atoms in all directions using the Linear Contraint Solver (LINCS) algorithm for bond constrains (Hess et al. 1997). Smooth particle mesh Eldward (PME) algorithm was used to estimate the electrostatic interactions (Darden et al. 1993). The cut-off range of electrostatic and Van-der-Waals interactions was set to 1.6 Å for both. The H-bonds formed among ligands, receptors, and solvent molecules were analysed using the built-in hbond modules in GROMACS, with a cut-off radius of 35 Å and angle of 30°. The binding affinities for the last 5 ns of the simulation were evaluated using gmx_MMPBSA (Valdés-Tresanco et al. 2021) for each complex. The root mean square error (RMSD) was evaluated along all the frames with GROMACS.
In Vivo Hepatic Exposure
Rat (Rattus norvegicus) hepatic gene expression data from in vivo exposure were obtained from the Open Toxicogenomics Project-Genomics Assisted Toxicity Evaluation System (TG-GATEs) and Gene Expression Omnibus (GEO) datasets (www.ncbi.nlm.nih.gov/gds): The Open TG-GATEs EE2 in vivo treatment was described previously (Igarashi et al. 2015). Data from 6- and 9-h exposures to 3 mg/kg EE2 were chosen for downstream analysis. AFB1 in vivo sample datasets were obtained from NCBI GEO (Accession:GSE57815) (Gusenleitner et al. 2014); these samples represented 3 and 5-day AFB1 exposures respectively to 0.3 mg/kg bw × day. Considering the average weight of male Sprague Dawley rat is comprised between 450 and 520 g (Sprague Dawley), the average AFB1 dosage was of 145.5 ± 14.8 μg/day. This value is in the range of the average maximum occurrence of AFB1 in food commodities reported over the last ten years (108 ± 182.5 μg/kg) (Rushing and Selim 2019).
Liver Differential Gene Expression Analysis
All CEL files obtained from public databases were analyzed using R (version 3.6.1) (R Core Team 2017), R studio (version 1.1.463) (RStudio Team 2020) and packages present in Bioconductor (version 3.11) (Gentleman et al. 2004; Huber et al. 2015). CEL file upload, pre-processing and normalization was done using affy (version 1.66.0) (Gautier et al. 2004), Gene expression analysis was carried out using Limma (version 3.44.3) (Ritchie et al. 2015) and Biobase (version 2.48.0) (Huber et al. 2015) from Bioconductor (www.bioconductor.org). Rat in vivo samples were analyzed with the Affymetrix Rat Genome 230 2.0 Array for both exposures (AFB1 and EE2). Probe ID to Gene Symbol conversions were obtained from the ThermoFisher (Affymetrix) website (www.thermofisher.com). Rat in vivo microarray data, before being processed using Bioconductor packages, were appended with human gene IDs using the orthology search (g:Orth) function on g:Profiler (Raudvere et al. 2019) to link probe IDs to human gene symbols. This resulted in fewer gene symbols in the limma output, but as human gene annotation is richer in terms of genomic annotation this was selected to enable a more accurate and human centric downstream analyses including gene ontology (GO) and pathway level analysis. To select a biologically relevant number of Differentially Expressed (DE) genes and keep the ranked lists statistically significant the outputs were filtered by three different settings of adj. p-value, or q-value (0.05-very stringent, 0.1-stringent and 0.4-modest, respectively). The q-value corresponds to the Benjamini and Hochberg (1995) correction of p value or False Discovery Rate (FDR).
Gene Ontology (GO) Analysis
GO Biological Process (BP) enrichment and meta-analysis of differential gene expression output from limma was analyzed using iPathwayGuide (Advaita Bioinformatics) (Ahsan and Drăghici 2017). To obtain enough differentially expressed genes suitable for systems-level analysis, a threshold q-value of 0.4 of was applied. This q-value cut off was guided by the literature and previous articles on the study of estrogen receptor response (Dhamad et al. 2016; Hardiman et al. 2016; Huff et al. 2018, 2019; Irish et al. 2016).
Network Construction
With the aim of carrying out contextual PPI network analysis, we pursued identification of the most central nodes and subsequently compared these with known estrogen-responsive targets (ERT). To highlight ER involvement in modulating gene expression from AFB1 exposures, PPI networks were constructed. The gene lists obtained from the AFB1 treatments used in the GO analysis (in vivo AFB1 0.3 mg/kg bw × day: genes filtered by a q-value threshold of 0.4) were added with estrogen receptors and G coupled-protein estrogen receptor 1 gene symbols (ESR1, ESR2 and GPER1) to better evaluate the estrogenic component of the network. The gene lists were inputted to the String Database (Szklarczyk et al. 2019) to construct PPI networks based on a "combined score" deriving from text-mining, experiments, curated databases (e.g. KEGG), co-expression, neighborhood, gene fusion and co-occurrence evidence. The networks obtained were subsequently imported into Cytoscape (version 3.8.0) (Shannon et al. 2003) for downstream analysis.
Estrogen Responsive Targets
The list of Estrogen Responsive Targets (ERT) was obtained using the "consensome" function from Ominer (signalingpathways.org) (Ochsner et al. 2019). To facilitate comparison with the proteins in the PPI networks constructed from the AFB1 exposure data generated using STRING, the ERT list was inputted in g:Convert of g:Profiler to convert the HGNC terms to Ensemble protein IDs (ENSPs) which resulted in a list of 12,365 redundant ENSPs. For every network we considered only the ERT effectively present in the network, this step removed redundancy and focused the attention only on gene expression resulting from AFB1 exposures.
Network Analysis
Essential proteins are those whose removal can cause lethal defects (Lei et al. 2019). To highlight the most important nodes in the networks the procedure of identification of essential proteins previously described was modified (Tang et al. 2015). The list of ERTs was inputted into the network analysis, allowing comparison of estrogen receptor targets with the core nodes of the network.
Oxidative Stress Response in the Core Networks
As AFB1 acts via increases in oxidative stress, we checked the involvement of ROS and oxidative stress in the core networks. The list of nodes from the core networks at 3- and 5-days were inputted into ToppFun (Chen et al. 2009) to facilitate GO enrichment analysis, the B&H q-value threshold was set at 0.05. From the list of enriched BPs, we selected the terms and genes inherent to “oxidative stress” and “reactive oxygen species”. Finally, we calculated the ratios between the BP and genes to evaluate the involvement of the oxidative stress response (Eq. 1).
Equation 1. Ratios used to evaluate the oxidative stress response in the core networks.
Graphics and figures were generated using the Moderndive R package (version 0.4.0) (Ismay and Kim 2019), the GNU Image Manipulator Program (version 2.10.18) (The GIMP Development Team 2019), ADT (version 1.5.6) and Cytoscape (version 3.8.0).
In-Depth Information
More detailed information is available in supplemental methods.
Results
Molecular Docking Simulations
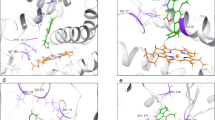
AutoDock4 simulations on ERα revealed the high affinity of EE2 for the receptor. It was the best performing ligand with a binding affinity of − 10.98 kcal/mol and a KD of 10 nM. ZEA also an estrogenic control revealed a binding affinity of − 9.6 kcal/mol, and a KD of 90 nM. It was the second ranked compound in terms of its affinity for this receptor. Aflatoxins showed reduced affinities compared to both positive controls (AFB1: − 7.99, AFL: − 7.45, Epo-AFB1: − 7.82, AFM1: − 7.72 and AFQ1: − 8.06) and the KD s were in the micromolar range (μM). The two aflatoxins that bound the receptor with the best affinities were AFB1 and AFQ1 with KD s of 1.38 and 1.25 μM, respectively. Despite lower affinity values than the estrogenic positive controls, these data nevertheless suggest that AFB1 and its metabolites possess good affinity for ERα and can interact with the receptor at low concentrations. The binding energy of RIF, the selected negative control (340.71 kcal/mol) made unlikely any interaction between it and ERα (Table 1). All the ligands entered the ERα binding pocket. Focusing attention on oxygens and positioning of the aromatic rings, we observed that AFB1, AFL, AFM1 and Epo-AFB1 shared similar binding poses. AFQ1 had a pose more like EE2 than the other aflatoxins, especially the two six-carbon rings and O5 of AFQ1 which appeared in a similar position to the first aromatic ring, the second ring and O1 of EE2. Interestingly ZEA and EE2, the two positive controls do not appear to bind the receptor similarly, only O1 and O4 of ZEA showed positions close to O1 of EE2 (Fig. 2A).
Binding poses AutoDock4 simulations, obtained with AutoDock Tools (ADT). A Binding poses on ERα after AutoDock4 simulations, obtained with AutoDock Tools (ADT). The 11th α-helix is hidden to present a clearer visualization of the poses. This demonstrates that every ligand entered the binding site. Aflatoxins showed a similar binding pose among them excluding AFQ1, which had higher affinity for ERα and resulted in a pose more like EE2. Despite the high affinity of both EE2 and ZEA for ERα they do not share a similar binding pose. RIF, the negative control, entered the binding site, suggesting ligand low affinity for ERα. B Binding poses on ERβ after AutoDock4 simulations, obtained with ADT. The 10th α-helix was hidden to provide clearer visualization of the poses. All the ligands entered the ERβ binding site; aflatoxins result for the poses were less conserved than with ERα. AFB1 and AFM1 share similar binding poses, Epo-AFB1 and AFQ1 aromatic rings appear to be positioned in the receptor binding pocket in a similar way. ZEA resulted in a different pose to EE2. RIF entered the binding site, suggesting low affinity for the receptor
AutoDock4 simulations on the ERβ showed high affinities for the positive controls (EE2: − 9.72, ZEA: − 7.7 kcal/mol). The EE2 affinity and the resultant KD of 70 nM suggested high activity at low hormone concentrations. ZEA, despite its high affinity for ERα, had an affinity for ERβ of − 7.7 kcal/mol and a KD of 2.26 μM. These values are lower than those observed with AFB1 and AFQ1. Also, for AFB1, AFQ1 and AFL, the affinities for ERβ were slightly better than for ERα. The AFB1 binding affinity of − 8.1 kcal/mol and KD of 1.16 μM suggested activity at concentrations at lower levels than for ZEA. AFQ1 with an affinity of − 8.24 kcal/mol was the only aflatoxin with an estimated KD lower than 1 μM (0.9 μM). Considering the molar mass of AFQ1 (328.27 g/mol), the KD results in 295.4 μg/L of toxin, relatable to high levels of contamination (Rushing and Selim 2019). This result provides another indication of an inherent estrogenic component in aflatoxin contamination. The principal genotoxic metabolite of AFB1, Epo-AFB1, also resulted in a binding affinity better than ZEA (− 7.83 kcal/mol, KD: 1.81 μM). Other aflatoxins exhibited lower affinities (AFL: − 7.53, AFM1: − 7.54) than ZEA and KDs higher than 2.9 μM. RIF included as a negative control again yielded an unfavorable binding energy and no possibility of interacting effectively with the binding pocket of ERβ (Table 1). Binding poses in the ERβ binding pocket were less conserved among the aflatoxins. AFB1 and AFM1 share a similar binding pose. Additionally, the aromatic rings of AFQ1 appear positioned close to the Epo-AFB1 rings. Observing the aromatic ring of EE2, similarity was found close to the first aromatic ring of AFB1 and AFM1. O4 of both the aflatoxins places close to the O1 of EE2, and finally O2 of AFB1 and O5 of AFM1 are close to the O2 of EE2 (Fig. 2B). Similarly, to the ERα results, ZEA showed a different binding pose to EE2 except for the O2 of ZEA and O1 of EE2. A high degree of freedom is observed with ZEA due to the smaller number of intramolecular rings and sp2 hybridized carbons. RIF enters the binding site but shows low affinity for the receptor.
Molecular Dynamics Simulations
The calculated root mean square deviation (RMSD) for each ligand with ERα showed good stability for all the compounds. No ligand exceeded 0.5 Å of deviation (Fig. 3A, Table 2). In addition, the RMSD remained in a constant range along with all the frames suggesting the ligand’s stability in the binding pocket with time (Fig. 3A, Table 2). The obtained average and standard deviation RMSD values were: AFB1 0.385 ± 0.087 Å, AFQ1 0.418 ± 0.084 Å, EE2 0.281 ± 0.0747 Å, respectively. The calculated RMSD for each ligand with ERβ showed similar results, only AFB1 reached 0.5 Å of deviation and the RMSD remained within similar ranges along all frames (Fig. 3B, Table 2). Differently to ERα, the ligand with higher RMSD and standard deviation was AFB1: 0.530 ± 0.126 Å. The other ligands, AFQ1: 0.362 ± 0.058 Å, EE2 0.390 ± 0.075 Å, were more stable and constant along the time (Fig. 3B, Table 2).
Calculated RMSD among ligands and ERs. A AFB1, AFQ1 and EE2 RMSD calculated along the simulation time with the ERα. All the ligands showed a low and constant RMSD (< 0.33 Å) suggesting their stability into the binding pocket. B AFB1, AFQ1 and EE2 RMSD calculated along the simulation time with the ERβ. All the ligands showed a low and constant RMSD (< 0.39 Å) suggesting their stability into the binding pocket
Interestingly, from the H-bonds analysis among ligands and ERα, no H-bonds were formed between AFB1 and the receptor residues. Only up to 2 atom close pairs were identified along with the simulation. Instead, AFQ1 formed up to 2 contemporary H-bonds with the receptor residues, and they were present for 38.43% of the frames. Up to 4 atom close pairs were frequent along with the simulation. The synthetic estrogen EE2 formed up to 4 H-bonds with the receptor and they remained for almost all the simulation time (99.98%), the close pairs are up to 8 (Supplementary figures: Figure S6, Supplementary table: Table S1). With Erβ, AFB1, formed only one short-lasting H-bond. Moreover, the close pairs were more frequent during the first half of the simulation. In contrast to ERα, AFQ1 formed only 1 short-lasting H-bond with the Erβ residues. Nevertheless, the close pairs, up to 2, were constant along with all the frames. EE2 formed fewer H-bonds and close pairs with Erβ than with ERα, the maximum number of H-bonds was 3, and 5 close pairs. The H-bonds lasted for most of the frames (91.72%) (Supplementary figures: Figure S7, Supplementary table: Table S1).
As suggested by the previous analyses, gmx-MMPBSA estimated the best binding free energy with ERα for EE2 (− 48.5 ± 2.3 kcal/mol), the two toxins also showed a good affinity for the ERα, both in a similar range (AFB1: − 36.5 ± 2.7, AFQ1: − 35.2 ± 2.7 kcal/mol). The energies were stable along with the frames for every ligand, suggesting their stability in respect of the binding pocket (Fig. 4A, Table 3). The results for the ERβ are like the receptor α, EE2 showed the best free energy of binding (− 49.7 ± 2.9 kcal/mol), the two toxins also showed a good affinity for the ERβ, both in a similar range (AFB1: − 35.0 ± 2.2, AFQ1: − 36.0 ± 2.2 kcal/mol). The energies were almost constant along with the frames, with low fluctuation, suggesting the stability of all the ligands into the binding pocket (Fig. 4B, Table 3).
Calculated free energy of binding among ligands and ERs. A AFB1, AFQ1 and EE2 free energies of binding estimated along the simulation time with the ERα. As suggested by pervious analysis, EE2 showed the best free energy of binding ( − 48.5 ± 2.3 kcal/mol), the two toxins also showed a good affinity for the ERα, both in a similar range (AFB1: − 36.5 ± 2.7, AFQ1: − 35.2 ± 2.7 kcal/mol). The energies were stable along the frames for every ligand, suggesting the stability of them in respect of the binding pocket. B The results for the ERβ are similar to the receptor α, EE2 showed the best free energy of binding (− 49.7 ± 2.9 kcal/mol), the two toxins also showed a good affinity for the ERβ, both in a similar range (AFB1: − 35.0 ± 2.2, AFQ1: − 36.0 ± 2.2 kcal/mol). The energies were stable along the frames, with lower fluctuation (sd) if compared with the energies estimated with the ERα
Liver Differential Gene Expression Analyses
To obtain sufficient DE genes for GO and system-level analysis the modest 0.4 q-value threshold was selected yielding 1456 genes for the 3-day exposure and 605 genes for the 5-day exposure (Table 4). The EE2 exposures showed a higher numbers of significant DE genes, however the same q-value threshold used with AFB1 exposure (i.e., 0.4) was adopted for the EE2 exposures (Table 4).
To compare the gene expression patterns shared by the different exposures a Venn Diagram was generated (Fig. 5A and Table 5). Comparison within DE genes from the same exposure revealed partial overlap. The 3- and 5-day AFB1 samples shared only 106 DE genes out of a total of 1897. Also, the unique signatures of the two different time points differed, 852 genes for the 3-day exposure and 333 genes for the 5-day exposure. The EE2 exposure at 6 and 9 h showed partial overlap; of the 3653 DE genes obtained from both EE2 exposures, only 1212 were shared by both time points. Moreover, 997 of these 1212 DE genes were not present in the AFB1 expression signature. Similarly to AFB1, the EE2 unique signatures differed; only 372 genes were expressed after EE2 exposure for 6 h and 1636 for 9 h (Fig. 5A and Table 5).
Venn Diagrams showing overlaps of differentially expressed (DE) genes and enriched Gene Ontology Biological Process Terms (BP) among AFB1 (0.3 mg/kg bw × day exposure after 3 and 5-day exposures) and EE2 (3 mg/kg exposure after 6 and 9 h). A DE genes in the AFB1 and EE2 exposures. Genes differentially expressed after AFB1 and EE2 exposures showed a partial overlap. Interestingly a comparison within the same compound also showed a partial overlap, and for AFB1 exposures was lower than among AFB1 and EE2. B BP observed after GO analysis for AFB1 and EE2 exposures. The BP also resulted in a partial overlap in enriched terms among the AFB1 and EE2 exposures. In addition, as seen in expression analysis there is higher overlap among AFB1 3-day and both the EE2 exposures
Comparison between the two compounds revealed that AFB1 at 3 and 5-days differed in the number of DE genes that were shared with the EE2 exposures. The 3-day exposure to AFB1 shared 243 DE genes with EE2 expression after 6 h and 450 genes with EE2 expression after 9 h of exposure. The AFB1 5-day exposure shared 89 DE genes in common with the 6 h EE2 exposure and 161 genes with the 9 h EE2 exposure (Fig. 5A and Table 5).The 5-day AFB1 exposure shared fewer genes with AFB1 at 3-day than with the EE2 exposures.
Gene Ontology Enrichment Analysis
To compare biological effects among the exposures, the limma outputs filtered by q-values < 0.4 were inputted into iPathwayGuide (AdvaitaBio). Enriched biological process (BP) terms revealed a partial overlap between the AFB1 and EE2 exposures. The EE2 exposures showed strong unique signatures of 157 and 202 BP terms respectively for the 6 and 9-h exposures. Furthermore, a significant number of BP terms was shared by both exposures (134); of these 102 were not related to AFB1 exposures. Interestingly the AFB1 3 and 5-day exposures shared only 4 BP terms, and of these only 2 were not related to EE2 exposure. The aflatoxin unique signatures were 146 enriched BP terms for the 3-day exposure and 181 for the 5-day exposure. Moreover, as seen with the gene level analysis there are higher overlapping BP terms among the AFB1 3-day and EE2 exposures than with AFB1 at 5-days of exposure, resulting in a total of 37 shared BP terms. Of these BP terms shared by the AFB1 3-day exposure, 27 overlap with both EE2 exposures. The AFB1 3-day exposure shared 5 additional BP terms with both EE2 experiments. The AFB1 5-day exposure yielded 22 enriched BP terms shared with both EE2 exposures (Fig. 5B and Table 6).
AFB1 3-Day Exposure
As noted above the AFB1 3-day exposure and enriched GO terms had more in common with EE2 than with the AFB1 5-day exposure. Of the 27 GO terms present in both EE2 exposures strong correlations with immune and deregulation of inflammation was found (seven BP terms). Other enriched BP terms included protein regulation, including ubiquitination, protein targeting and folding. Of the 5 BP terms exclusively shared by the 3-day AFB1 and 6-h EE2 exposure, three were related to metabolic processes, one with energy metabolism (positive regulation of cristae formation, q-value: 0.012) and one differentiation related process (postsynaptic actin cytoskeleton organization q-value: 0.032). BP terms unique for the AFB1 3-day and EE2 9 h data were related to DNA damage (positive regulation of double-strand break repair: q-value 0.032, response to X-ray q-value: 0.014) and inflammatory conditions (complement activation, alternative pathway q-value: 0.024). The complete list of BP terms shared by EE2 exposures and AFB1 3-day exposure is in Table 7.
AFB1 5-Day Exposure
The 5-day AFB1 experiment resulted in 22 enriched BP terms shared with the EE2 exposures, despite the lower number of terms than the AFB1 3-day exposure some are interesting for their involvement in estrogenic response and cancer development. Indeed, two processes concerning telomerase activity were deregulated in the AFB1 and both EE2 exposures (positive regulation of telomerase activity q-value: 0.039 and positive regulation of telomere maintenance via telomerase q-value: 0.005). Other processes involving protein regulation and metabolism were present in both the EE2 exposures. The 9-h EE2 and 5-day AFB1 exposures shared additional key processes. Two enriched BPs are in response to gamma radiation (q-value = 0.01777) and X-rays (q-value = 0.02367) highlighting a DNA damage situation. Four additional BPs are involved in the innate (complement regulation: 0.00645 and activation: 0.0112) and adaptive (MHC II antigen presentation: 0.04688, T cell apoptosis regulation: 0.00101) immune response. (Table 8). One more interesting term appears in the AFB1 5-day unique signature, with a q-value of 0.01, the BP cellular response to estrogen stimulus appears after only the 5-day AFB1 exposure and in none of the EE2 exposures (Supplementary Tables: Table S2). This BP term appears in the AFB1 5-day exposure because this is the only one exposure that led to a deregulation of ERα gene (ESR1) with a significant q-value (ESR1 Log2FC: − 0.864, q-value: 0.396).
PPI Network Analysis
AFB1 3-Day Exposure
The list of differentially expressed genes inherent to the AFB1 exposure, used in GO analysis (filtered q-value of 0.4), was inputted in STRING along with the ERs to construct a network consisting of 1390 nodes. Of these nodes, 242 were Estrogen Responsive Targets (ERT). Betweenness centrality results were scattered and able to link singular branches to the core network. Subgraph and closeness centralities resulted in more groups in the core of the network (Supplementary Figure S8). ERTs found among the most central nodes were variable, the betweenness centrality highlighted 56 ERTs, closeness centrality 64, and subgraph centrality 48. The Pearson’s chi-square test for independence rejected the null hypothesis indicating independence among the ERTs found in the entire network, and the ERTs recognized by single-centrality measures (χ2: 13.411, df: 3, p-value: 0.003828). Due to the differences among the centrality measures, the post-hoc FDR correction and pairwise testing demonstrated that ERTs found by betweenness, and closeness centrality were statistically significant (q-values; betweenness: 0.1242, closeness: 0.0073, subgraph: 0.4612), suggesting that the ERTs in the network are distributed in those nodes that are closer to all other nodes and can impair the regulatory network. The subnetwork resulting from overlapping nodes consisted of 120 nodes, 27 are ERTs and, excluding SMURF1, all the other nodes are their direct neighbors, including the estrogen receptors α and β, showing high correlation of the core network with ERTs and their direct neighbors (Fig. 6). The Pearson’s chi-square test for independence and FDR correction resulted in a q-value of 0.2, suggesting a significantly different distribution of ERTs between the entire network and the core network from a system-level analysis. (χ2: 1.6225, df: 1, p-value: 0.2027, q-value: 0.2). Moreover, a large involvement of the estrogenic component in the core network was observed, the 22.5% of nodes are ERTs (27 nodes) and 76.7% are their direct neighbors (92 nodes).
AFB1 5-Day Exposure
The list of differentially expressed genes inherent to the AFB1 exposure, used in GO analysis (filtered q-value of 0.4), added with estrogen receptors genes, was inputted in STRING to construct a network consisting of 574 nodes, 101 of which are ERTs. Similarly, to previous analyses, betweenness centrality highlighted nodes in a scattered distribution, subgraph and closeness centralities showed many overlapping nodes (Supplementary figures: Figure S9). Low amount of ERTs were found by centrality measure (betweenness: 20, closeness: 22, subgraph: 22) and the Pearson’s chi-square test for independence did not reject the Null hypothesis thus not allowing any assumption regarding the distribution of the ERTs in the network (χ2: 1.7661, df: 3, p-value: 0.6223). Coherently, the post hoc FDR correction and pairwise testing showed that none of the independence of the ERTs found by the centrality measure were statistically significative (q-value; betweenness: 1, closeness: 1, subgraph: 1). The overlapping nodes produced a subnetwork of 43 nodes, of which only 6 are ERTs and 21 their direct neighbors. There are 16 nodes in the subnetwork that are not involved in the estrogenic component and suggest a lower involvement of ER pathways in the 5-day exposure to AFB1. Despite the lower correlation with estrogenic components both ERα and β are present in the subnetwork (Fig. 7). The Pearson’s chi-square test for independence and FDR correction did not reject the null hypothesis thus not allowing the assertion that the ERTs found in the subnetwork are independent of the ERTs distributed in the entire network (χ2: 0.15973, df: 1, p-value: 0.6894, q-value: 0.69).
Most central nodes and ERTs of the PPI network from AFB1 in vivo 5-day exposure. Lower correlation with estrogenic components were found, only 6 proteins were ERTs with 21 direct neighbors. 16 nodes do not share direct contacts with ERTs. Despite the lower degree of estrogenic correlation both estrogen receptors were found in the subnetwork
Core Networks Oxidative Stress Involvement
The 120 nodes from the 3-day core networks resulted in 1389 significantly enriched BP terms. From these 20 terms were inherent to the oxidative stress response, with a total of 19 related nodes. 1.8% of BP terms and 15.8% of nodes were inherent to oxidative stress. 43 nodes from the 5-day core networks yielded 1176 significantly enriched BP terms. From these 20 BP terms were inherent to the oxidative stress response, with a total of 12 related nodes. 1.7% of BP terms and the 27.9% of nodes were inherent to oxidative stress.
Discussion
Different observations can be drawn from the molecular docking and molecular dynamics simulations. The free energies of binding and estimated KD obtained demonstrate that AFB1 and its metabolite AFQ1 can interact with the estrogen receptors α and β (Cotterill et al. 2019). AFQ1 exhibited greater affinity for ERβ than the mycoestrogen ZEA (Table 1). Moreover, the constants of dissociation at a micromolar (μM) level, although not typical for high-affinity ligands, are used in chemical gene interaction studies (Reddy et al. 2004) and to highlight structures that can interact, and even inhibit, specific receptors and transcription factors (Koehler et al. 2003). This suggests a mechanism of endocrine disruption for AFB1 and its metabolites, resulting from repeated intake of contaminated foods, targeted against the ERs, and resulting in an estrogenic signaling cascade. Interactions formed among the analyzed compounds and residues in the binding pocket of the ERs revealed similarities with the positive estrogenic control EE2. AFB1, some of its metabolites and EE2 formed a H-Bond with the same residue (Arg394) when binding ERα and shared several weak interacting residues. Additionally, the interaction between 17-β-Estradiol (EST) and ERα in the PDB crystallized structure on PDB (1ERE) (Supplementary Figure S10), revealed that in addition to the same H-Bond previously highlighted, another specific interaction was shared among EST, AFB1 and all its metabolites. Phe404 forms stacking interactions with the aromatic ring of the ligands (Supplementary Figure S10). When we superimposed ERα and β we uncovered Phe356 in the ERβ binding pocket in the same position of Phe404 in ERα suggesting a similar mechanism for the ERs in binding ligands (Supplementary Figure S10). This residue forms the same stacking interactions with AFB1, all its metabolites and EE2 from the docking simulations (Supplementary Figures S2–S5). Interestingly the different H-Bonds formed among ERs residues and the ligand, besides several weak interactions shared with EE2, suggests a differential effect of the ligand on ERs, similar to ZEA that despite high affinity does not form a H-Bond with ERs residues (Supplementary Figures S3, S5). Taken together these results suggest that AFB1 and its metabolites can disrupt the activity of ERs, exhibiting high binding affinity and, excluding AFQ1, utilize similar binding modes as with the positive control EE2. The differences in binding position also suggest the different effects of aflatoxins on the activation of ERs compared to natural and synthetic activators such as EST and EE2. Taken together one can speculate that AFB1 and AFQ1 could bind at similar concentrations to ERα but ultimately leading to different activation states due to the different interaction specificity.
Differential gene expression and overlap among the EE2 3 mg/kg 6 and 9-h treatments and AFB1 0.3 mg/kg bw × day treatments suggest the possibility of a common mechanism of action. The higher number of deregulated genes among the AFB1 3-day and EE2 treatments and AFB1 5-day and EE2 9-h treatments than those genes shared by the two AFB1 treatments suggests the existence of an estrogenic component to the mechanism of action of Aflatoxin. Moreover, the lower number of deregulated genes shared by the 5-day treatment with AFB1 and EE2 suggests that the accumulation of cytotoxic effects caused by AFB1 exceed the estrogenic effects found with the 3-day AFB1 exposure. In the BP chaperone-mediated protein folding term (q-value: 0.00844) the heat-shock protein HSPA8 was upregulated by AFB1 3-day treatment and both EE2 treatments, only in the AFB1 5-day treatment it resulted in down-regulation (log2FC; AFB1 3-day: 1.180, AFB1 5-day: − 0.396, EE2 6 h: 0.996, EE2 9 h: 1.308). Interestingly this, and other heat-shock proteins are directly related to ERα activity (Dhamad et al. 2016), the different level of expression seen with the AFB1 5-day treatment strengthens the hypothesis that a longer exposure to AFB1 results in more cytotoxic effects than ER related activities. Also, the up-regulation of transferrin transport found after the 5-day AFB1 treatment (q-value: 0.0478) appears to be linked to oral contraceptive intake (Bhagavan and Ha 2015).
Evidence suggests that NF-κB responds to estrogen modulation through the activity of ERs (Xing et al. 2012). Moreover, deregulation of the non-canonical NF-κB pathway and NFκB-inducing kinase (NIK) expression was found in cancer (Demchenko et al. 2010) and appears to be regulated by estrogenic activities (Ansari and Gandy 2007; Biswas et al. 2005). Observing deregulated genes involved in the BP term inherent to the non-canonical NF-κB signaling pathway NIK/NF-kappaB signaling (q-value: 0.02581), we noted that the AKT serine/threonine kinase 1 ATK1 was up-regulated after 3-day of AFB1 treatment and in both the EE2 treatments at similar levels (log2FC; AFB1 3-day: 0.515, EE2 6 h: 0.470, EE2 9 h: 0.510). AKT1 is an estrogen activated gene, likely via non-nuclear estrogen receptors such as GPER1 (Rogers et al. 2009) and involved in suppression of apoptosis through NF-κB non-canonical signaling (Madrid et al. 2000). This indicates that the proliferative effects of estradiol could be also shared by the early exposure response to AFB1. DNA replication-dependent nucleosome assembly (q-value: 0.04657) is another term that is involved in proliferation and found in the AFB1 3-day and both EE2 treatments. Here the gene encoding the chromatin remodeling factor RB binding protein 4 (RBBP4) was up-regulated in all treatments (log2FC; AFB1 3-day: 0.832, AFB1 5-day: 0.275, EE2 6 h: 0.491, EE2 9 h: 0.343). Nevertheless, RBBP4 in the 5-day AFB1 exposure was not statistically significant (q-value: 0.49). This protein is reported to interact with ERα and to modulate the estrogen-responsiveness in MCF-7 cells (Creekmore et al. 2008). RBBP4 interacts directly with the tumor suppressor RB and its expression is up-regulated in human primary HCC (Song 2004). Moreover, this gene is reported to be a target of NF-κB. Inhibiting NF-κB down-regulated RBBP4 suggesting its involvement in ER related pathways and proliferative effects (Pacifico et al. 2007). Looking at other deregulated genes involved in NF-κB signaling, in the BP term positive regulation of double-strand break repair (q-value: 0.03176), the gene MGMT (Guo et al. 2015) was found be differentially expressed in both the AFB1 and EE2 treatments relative to the controls. Indeed, MGMT encodes methylguanine methyltransferase which is involved in DNA repair from the damaging activity of alkylating agents. Furthermore, the genotoxic mechanism of action of AFB1 and its metabolites involves guanine alkylation (Benkerroum 2020). The differential expression of MGMT from the AFB1 3-day exposure resulted in a strong down-regulation (log2FC: − 2.207) while the longer exposure led to a strong up-regulation of the gene (log2FC; 2.467). EE2 exposures led to a lower but constant up-regulation of the gene (log2FC; 6 h: 0.553, 9 h: 0.866). ERα has been recognized as a regulator of the Wnt oncogene (Zhan et al. 2017), here we found 3 enriched BP terms relating to the deregulation of the Wnt pathway: Negative and positive regulation of canonical Wnt signaling (q-values: 0.02684 and 0.3642 respectively) and the Wnt signaling pathway and planar cell polarity pathway (q-value 0.0098).
These findings allow two interpretations. On one hand the downregulation found in the early AFB1 response suggests that the lower level of genotoxic effects in rats arose with the accumulation of AFB1 and its principal genotoxic metabolite Epo-AFB1. On the other hand, the strong initial downregulation suggests that the early AFB1 response resulted in a strong inhibition of DNA repair mechanisms that could enhance susceptibility to the genotoxic and cancerogenic effects of the mycotoxin. In addition, the higher expression of MGMT in EE2-treated rats was previously reported in vitro as a mechanism in response to ER-mediated DNA alkylation (Teo et al. 2001). A gene showing similar behavior to MGMT was NFE2L2, indeed from the AFB1 exposures it was initially down-regulated and subsequently up-regulated (log2FC; AFB1 3-day: − 1.220, AFB1 5-day: 0.628 log2FC). Similarly, to MGMT, NFE2L2 was slightly upregulated by the EE2 treatments (log2FC; EE2 6 h: 0.593, EE2 9 h: 0.767). NFE2L2 encodes a transcription factor that regulates genes containing antioxidant response elements in response to oxidative stress, its downregulation in the initial phase of AFB1 treatment suggests a lower level of oxidative stress and DNA damage compared with longer duration AFB1 exposures or an increased susceptibility to oxidative stress related DNA damage. E2 and ER related pathways were previously associated with the increase of oxidative stress and genotoxic effects (Lam et al. 2011; Roy et al. 2007). These effects explain the upregulation of NFE2L2 expression. The differential expression of MGMT and NFE2L2 were not directly linked with our hypothesis of estrogenic components in the AFB1 mechanism of action, but hint that the initial exposure to AFB1 could inhibit different and unrelated mechanisms of protection for DNA damage, improving the susceptibility and deleterious effects that the toxin exerts against hepatocytes.
Another differentially expressed gene that showed effects similar to that previously reported in the literature is TP53, from which is already known that the genotoxic effects of AFB1 are particularly focused on codon 249 which could compromise normal expression of this tumor suppressor gene (Benkerroum 2020; EFSA Panel on Contaminants in the Food Chain (CONTAM) et al. 2020). In our analysis, TP53 was not deregulated in the early exposure to AFB1 but after 5 days of treatment there was a significant down-regulation of the gene (log2FC; − 0.510). On the other hand, EE2 slightly up-regulated TP53 in both treatments (log2FC; EE2 6 h: 0.399, EE2 9 h: 0.378). These results suggest that the genotoxic effects of AFB1 against TP53 are mostly like to rise with a longer exposure, leading to a down-regulation of the gene and subsequent oncogenic effects. Finally, the presence of the BP cellular response to estrogen only after the 5-day treatment with AFB1 appears controversial. It could suggest the involvement of estrogenic pathways in AFB1 exposure, but it appears in none of the EE2 exposures.
AFB1 0.3 mg/kg bw × day treatment for 3 days led to a core network of 120 nodes, 22.5% of these are ERTs (27 nodes) and 76.7% are direct neighbors (92 nodes). 99.2% of the core network is related to estrogenic targets, and of these nodes some key proteins highlighted in the expression analysis were revealed to play a crucial role in regulatory process, these proteins include AKT1, RBBP4 and HSPA8. Moreover, many nodes inherent to translation such as the elongation factors EIF4A1, EIF4G1 and many ribosomal proteins including RPS5, RPS7, RPS15 suggest a strong impairment of protein synthesis, already known as an AFB1 exposure effect (Benkerroum 2020). In addition, numerous nodes inherent to proteasomal sub-units including PSMA1, PSMA5, PSMC7, PSMD6 suggests two things, the first is deregulation of protein stability, in line with previous knowledge on AFB1 effects, and the second is the possible deregulation of cell cycle due to the involvement of the proteasome in cell cycle regulation (Adams 2004). In support of the deregulation of cell cycle other key factors are found as core nodes, the cell division cycle proteins CDC23 and CDC42 that directly interact with ERβ (Giurato et al. 2018), the cyclin dependent kinase 2 (CDK2) that was shown to be down-regulated by long-term estradiol treatments and to prevent liver regeneration (Foster and Wimalasena 1996) and finally calmondulin3 (CALM3), a regulator of the cell cycle and cytokinesis whose promoter contains estrogen responsive elements and can modulate ERα activity, stabilizing the receptor dimers and increasing its activity (Li et al. 2017; Pedrero et al. 2002).
The PPI network from the 5-day AFB1 treatment resulted in a subnetwork smaller and less related to estrogenic targets, of the 43 nodes only 6 were ERTs (14%) and 21 their direct neighbors (48.8%) suggesting a significant lower involvement of the estrogenic components in the network and expression regulation. Observing the most central nodes, we noted TP53, suggesting a high involvement of cytotoxic and genotoxic effects leading to apoptotic processes. A further indication of the accumulation of genotoxic effects is the presence of NFE2L2, whose expression was up-regulated after 5-days of treatment and suggests an increased level of oxidative stress due to accumulation of AFB1 metabolites such as Epo-AFB1. Another interesting node is the proto-oncogene MDM2, after 5-days of exposure its expression was increased in the rat liver (log2FC; 2.094). Moreover, the presence of CDK9 and the transforming growth factor alpha TGFA, an ERT, highlight the strong tumorigenic component of the core network, suggesting that prolonged exposure to AFB1 and its metabolites, and the accumulation of genotoxic effects resulted in carcinogenic development by the direct DNA damage exerted by the toxins. Analysis of the oxidative stress showed a minor role in core networks. Like the network analysis, the 5-day treatment showed less estrogenic involvement and higher oxidative stress than the 3-day treatment.
Conclusions
In this paper, we present an innovative approach linking system biology and computational toxicology to elucidate the MOA of food toxicants. The integration of different bioinformatics and computational biology approaches uncovered the ability of Aflatoxins to interact with ERs, and with better affinities for ERβ than zearalenone. An overlap in differentially expressed genes and biological processes among AFB1 and EE2 exposures in vivo, highlighted key biological processes involved in the progression of liver disease and carcinogenesis, the principal effect of aflatoxin on the liver. Finally, strong estrogenic components at the core of PPI networks constructed using gene expression information, provided important clues to the role of estrogenic signaling in the MOA of aflatoxin. All these results indicate that the AFB1 is a potential endocrine disruptor, with the potential to alter the activation state of nuclear and membrane hepatic ERs. These results are strengthened by the ambiguous effect of estradiol and EE2 on the liver, indeed a physiological exposure to estradiol appears to protect against the development of HCC (Palmisano et al. 2017) but estrogen deregulation and exposure to xenoestrogens increase the chance of developing HCC (Notch et al. 2007; Wan and O’Brien 2014). The study presents some limitations including the use of only transcriptomics data, a small sample size and a limited range of concentrations tested, with expression data derived from different sources. Including more samples, newer technologies such as high throughput RNA sequencing and additional concentrations would address these limitations and will be the subject of future research. Also, as the study was computational in nature, in vitro binding tests across different concentration ranges will be required to confirm if aflatoxins bind effectively to human ERs. In vivo exposures with lower concentrations of both compounds will provide insights into the toxicological effects on hepatocytes at environmental and physiologically relevant concentrations. Overexpression and ablation of key genes found in our expression analysis will confirm the involvement in HCC development. In conclusion, this integrative systems approach has provided novel insights into the estrogenic component of the MOA of aflatoxins.
Data Availability
Enquiries about data availability should be directed to the authors.
References
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Adams J (2004) The proteasome in cell-cycle regulation. In: Adams J (ed) Proteasome inhibitors in cancer therapy. Humana Press, Totowa, pp 77–84
Ahsan S, Drăghici S (2017) Identifying significantly impacted pathways and putative mechanisms with iPathwayGuide. Curr Protocols Bioinf. https://doi.org/10.1002/cpbi.24
Ansari RA, Gandy J (2007) Determining the transrepression activity of xenoestrogen on nuclear factor-kappaB in Cos-1 cells by estrogen receptor-a. Int J Toxicol 26:441–449. https://doi.org/10.1080/10915810701620317
Battilani P, Toscano P, der Fels-Klerx HJV, Moretti A, Leggieri MC, Brera C et al (2016) Aflatoxin B1 contamination in maize in Europe increases due to climate change. Sci Rep. https://doi.org/10.1038/srep24328
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (methodol) 57:289–300
Benkerroum N (2020) Chronic and acute toxicities of aflatoxins: mechanisms of action. Int J Environ Res Public Health 17:423. https://doi.org/10.3390/ijerph17020423
Beshir S, Shaheen W, Saad-Hussein A, Saeed Y (2020) Aflatoxin B1 as an endocrine disruptor among miller flour workers. South Eastern Eur J Public Health (SEEJPH) XIV:2020. https://doi.org/10.4119/SEEJPH-3441
Bhagavan NV, Ha C-E (2015) Metabolism of iron and heme. In: Bhagavan NV, Ha C-E (eds) Essentials of medical biochemistry, 2nd edn. Academic Press, San Diego, pp 511–529
Biswas DK, Singh S, Shi Q, Pardee AB, Iglehart JD (2005) Crossroads of estrogen receptor and NF-kappaB signaling. Sci Signal 2005:pe27. https://doi.org/10.1126/stke.2882005pe27
Blankenship LT, Dickey JF, Bodine AB (1982) Invitro mycotoxin binding to bovine uterine steroid hormone receptors. Theriogenology 17:325–331. https://doi.org/10.1016/0093-691X(82)90092-9
Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engström O et al (1997) Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389:753–758. https://doi.org/10.1038/39645
Casey JR, Grinstein S, Orlowski J (2010) Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol 11:50–61. https://doi.org/10.1038/nrm2820
CCCF14. 2020. CCCF14 2020 | CODEXALIMENTARIUS FAO-WHO. https://www.fao.org/fao-who-codexalimentarius/meetings/extra/cccf14-2020/en/. [accessed 23 Jan 2022].
Centre international de recherche sur le cancer, ed. (2002) Some traditional herbal medicines, some mycotoxins, naphtalene and styrene. Lyon, World Health Organization
Chen J, Bardes EE, Aronow BJ, Jegga AG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37:W305–W311
European Commission. 2006a. Commission Regulation (EC) No 401/20006a of 23 February 2006a laying down the methods of sampling and analysis for the official control of the levels of mycotoxins in foodstuffs. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:02006R0401-20140701 [accessed 23 Jan 2022].
European Commission. 2006b. Commission Regulation (EC) No 1881/2006b of 19 December 2006b setting maximum levels for certain contaminants in foodstuffs. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32006R1881 [accessed 23 Jan 2022].
Cotterill JV, Palazzolo L, Ridgway C, Price N, Rorije E, Moretto A et al (2019) Predicting estrogen receptor binding of chemicals using a suite of in silico methods—complementary approaches of (Q)SAR, molecular docking and molecular dynamics. Toxicol Appl Pharmacol 378:114630. https://doi.org/10.1016/j.taap.2019.114630
Creekmore AL, Walt KA, Schultz-Norton JR, Ziegler YS, McLeod IX, Yates JR et al (2008) The role of retinoblastoma-associated proteins 46 and 48 in estrogen receptor alpha mediated gene expression. Mol Cell Endocrinol 291:79–86. https://doi.org/10.1016/j.mce.2008.05.016
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM (2010) Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood 115:3541–3552. https://doi.org/10.1182/blood-2009-09-243535
Dhamad AE, Zhou Z, Zhou J, Du Y (2016) Systematic proteomic identification of the heat shock proteins (HSP) that interact with estrogen receptor alpha (ERalpha) and biochemical characterization of the ERalpha-Hsp70 interaction: D. Picard, ed. PLoS ONE 11:e0160312. https://doi.org/10.1371/journal.pone.0160312
EFSA Panel on Contaminants in the Food Chain (CONTAM), Schrenk D, Bignami M, Bodin L, Chipman JK, del Mazo J et al (2020) Risk assessment of aflatoxins in food. EFS2. https://doi.org/10.2903/j.efsa.2020.6040
Eskola M, Kos G, Elliott CT, Hajšlová J, Mayar S, Krska R (2020) Worldwide contamination of food-crops with mycotoxins: validity of the widely cited ‘FAO estimate’ of 25%. Crit Rev Food Sci Nutr 60:2773–2789. https://doi.org/10.1080/10408398.2019.1658570
FDA. 2021. CPG Sec 570.375 Aflatoxins in peanuts and peanut products | FDA. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/cpg-sec-570375-aflatoxins-peanuts-and-peanut-products [accessed 23 Jan 2022].
Food safety digest, department of food safety and Zoonoses. World Health Organisation (WHO) Aflatoxins.
Foster JS, Wimalasena J (1996) Estrogen regulates activity of cyclyn-dependent kinases and retinoblastoma protein phosphorilation in beast cancer cells. Mol Endocrinol 10:488–498. https://doi.org/10.1210/mend.10.5.8732680
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315. https://doi.org/10.1093/bioinformatics/btg405
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. https://doi.org/10.1186/gb-2004-5-10-r80
Giurato G, Nassa G, Salvati A, Alexandrova E, Rizzo F, Nyman TA et al (2018) Quantitative mapping of RNA-mediated nuclear estrogen receptor beta interactome in human breast cancer cells. Sci Data. https://doi.org/10.1038/sdata.2018.31
Guo Y, Song T, Liu J, Li H, Tian Z, Xu C (2015) Disruption of NF-&kappaB signaling by fluoxetine attenuates MGMT expression in glioma cells. OncoTargets Therapy. https://doi.org/10.2147/ott.s85948
Gusenleitner D, Auerbach SS, Melia T, Gómez HF, Sherr DH, Monti S (2014) Genomic models of short-term exposure accurately predict long-term chemical carcinogenicity and identify putative mechanisms of action. PLoS ONE 9:e102579–e102579. https://doi.org/10.1371/journal.pone.0102579
Hardiman G, Savage SJ, Hazard ES, Wilson RC, Courtney SM, Smith MT et al (2016) Systems analysis of the prostate transcriptome in African-American men compared with European-American men. Pharmacogenomics 17:1129–1143. https://doi.org/10.2217/pgs-2016-0025
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: A linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472. https://doi.org/10.1002/(SICI)1096-987X(199709)18:12%3c1463::AID-JCC4%3e3.0.CO;2-H
Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS et al (2015) Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods 12:115–121. https://doi.org/10.1038/nmeth.3252
Huff M, da Silveira WA, Carnevali O, Renaud L, Hardiman G (2018) Systems analysis of the liver transcriptome in adult male zebrafish exposed to the plasticizer (2-Ethylhexyl) phthalate (DEHP). Sci Rep 8:2118. https://doi.org/10.1038/s41598-018-20266-8
Huff M, da Silveira W, Starr Hazard E, Courtney SM, Renaud L, Hardiman G (2019) Systems analysis of the liver transcriptome in adult male zebrafish exposed to the non-ionic surfactant nonylphenol. Gen Comp Endocrinol 271:1–14. https://doi.org/10.1016/j.ygcen.2018.10.016
Huuskonen P, Myllynen P, Storvik M, Pasanen M (2013) The effects of aflatoxin B1 on transporters and steroid metabolizing enzymes in JEG-3 cells. Toxicol Lett 218:200–206. https://doi.org/10.1016/j.toxlet.2013.01.015
Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T et al (2015) Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res 43:D921–D927. https://doi.org/10.1093/nar/gku955
Irish JC, Mills JN, Turner-Ivey B, Wilson RC, Guest ST, Rutkovsky A et al (2016) Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERα in SUM-44 breast cancer cells and is associated with ERα over-expression in breast cancer. Mol Oncol 10:850–865. https://doi.org/10.1016/j.molonc.2016.02.003
Ismay C, Kim A. 2019. Statistical inference via data science: a moderndive into R and the Tidyverse.
Koehler AN, Shamji AF, Schreiber SL (2003) Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. J Am Chem Soc 125:8420–8421. https://doi.org/10.1021/ja0352698
Kolawole O, Meneely J, Petchkongkaew A, Elliott C (2021) A review of mycotoxin biosynthetic pathways: associated genes and their expressions under the influence of climatic factors. Fungal Biol Rev 37:8–26. https://doi.org/10.1016/j.fbr.2021.04.003
Kourousekos GD, Theodosiadou EK (2015) Effects of aflatoxins on male reproductive system: a review. J Hellenic Vet Med Soc 66:201–210. https://doi.org/10.12681/jhvms.15863
Kourousekos GD, Theodosiadou EK, Lymberopoulos AG, Belibasaki S, Boscos C (2018) Effect of aflatoxin B1 on blood serum oestradiol-17β and progesterone concentrations during the luteal phase and the synchronized oestrus of goats. Anim Reprod 15:75–83. https://doi.org/10.21451/1984-3143-2017-AR939
Kyrein HJ (1974) The binding affinity of aflatoxins on the uterine oestrogen receptor. Z Lebensm Unters Forch 154:285–287. https://doi.org/10.1007/BF01083424
Lam SH, Lee SG, Lin CY, Thomsen JS, Fu PY, Murthy KR et al (2011) Molecular conservation of estrogen-response associated with cell cycle regulation, hormonal carcinogenesis and cancer in zebrafish and human cancer cell lines. BMC Med Genom. https://doi.org/10.1186/1755-8794-4-41
Lee Y, Lee J, Kim S, Lee S-C, Han J, Heu W et al (2014) Dissecting the critical factors for thermodynamic stability of modular proteins using molecular modeling approach. PLoS ONE 9:e98243. https://doi.org/10.1371/journal.pone.0098243
Lei X, Wang S, Wu F (2019) Identification of essential proteins based on improved HITS algorithm. Genes 10:177. https://doi.org/10.3390/genes10020177
Li Z, Zhang Y, Hedman AC, Ames JB, Sacks DB (2017) Calmodulin lobes facilitate dimerization and activation of estrogen receptor-alpha. J Biol Chem 292:4614–4622. https://doi.org/10.1074/jbc.m116.754804
Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO et al (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78:1950–1958. https://doi.org/10.1002/prot.22711
Madrid LV, Wang C-Y, Guttridge DC, Schottelius AJG, Baldwin AS, Mayo MW (2000) Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-kappaB. Mol Cell Biol 20:1626–1638. https://doi.org/10.1128/mcb.20.5.1626-1638.2000
Marchese S, Polo A, Ariano A, Velotto S, Costantini S, Severino L (2018) Aflatoxin B1 and M1: biological properties and their involvement in cancer development. Toxins 10:214. https://doi.org/10.3390/toxins10060214
Martínez-Rosell G, Giorgino T, De Fabritiis G (2017) Play molecule protein prepare: a web application for protein preparation for molecular dynamics simulations. J Chem Inf Model 57:1511–1516. https://doi.org/10.1021/acs.jcim.7b00190
Medina A, Rodriguez A, Magan N (2014) Effect of climate change on Aspergillus flavus and aflatoxin B1 production. Front Microbiol 5:348. https://doi.org/10.3389/fmicb.2014.00348
Mewshaw RE, Bowen SM, Harris HA, Xu ZB, Manas ES, Cohn ST (2007) ERb ligands, part 5: synthesis and structure–activity relationships of a series of 4-hydroxyphenyl-aryl-carbaldehyde oxime derivatives. Bioorg Med Chem Lett 17:902–906. https://doi.org/10.1016/j.bmcl.2006.11.066
Moore MM, Schoeny RS, Becker RA, White K, Pottenger LH (2018) Development of an adverse outcome pathway for chemically induced hepatocellular carcinoma: case study of AFB1, a human carcinogen with a mutagenic mode of action. Crit Rev Toxicol 48:312–337. https://doi.org/10.1080/10408444.2017.1423462
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Notch EG, Miniutti DM, Mayer GD (2007) 17alpha-Ethinylestradiol decreases expression of multiple hepatic nucleotide excision repair genes in zebrafish (Danio rerio). Aquat Toxicol 84:301–309. https://doi.org/10.1016/j.aquatox.2007.06.006
Ochsner SA, Abraham D, Martin K, Ding W, McOwiti A, Kankanamge W et al (2019) The signaling pathways project, an integrated omics knowledgebase for mammalian cellular signaling pathways. Scientific Data 6:252. https://doi.org/10.1038/s41597-019-0193-4
Pacifico F, Paolillo M, Chiappetta G, Crescenzi E, Arena S, Scaloni A et al (2007) RbAp48 is a target of nuclear factor-kappaB activity in thyroid cancer. J Clin Endocrinol Metab 92:1458–1466. https://doi.org/10.1210/jc.2006-2199
Páll S, Abraham MJ, Kutzner C, Hess B, Lindahl E (2015) Tackling exascale software challenges in molecular dynamics simulations with GROMACS. In: Markidis S, Laure E (eds) Solving software challenges for exascale. Lecture Notes in Computer Science. Springer International Publishing, Cham, pp 3–27
Palmisano BT, Zhu L, Stafford JM (2017) Role of estrogens in the regulation of liver lipid metabolism. In: Mauvais-Jarvis F (ed) Sex and gender factors affecting metabolic homeostasis, diabetes and obesity. Springer International Publishing, Cham, pp 227–256
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190. https://doi.org/10.1063/1.328693
Pedrero JMG, del Rio B, Martinez-Campa C, Muramatsu M, Lazo PS, Ramos S (2002) Calmodulin is a selective modulator of estrogen receptors. Mol Endocrinol 16:947–960. https://doi.org/10.1210/mend.16.5.0830
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC et al (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H et al (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47:W191–W198. https://doi.org/10.1093/nar/gkz369
Reddy MM, Bachhawat-Sikder K, Kodadek T (2004) Transformation of low-affinity lead compounds into high-affinity protein capture agents. Chem Biol 11:1127–1137. https://doi.org/10.1016/j.chembiol.2004.05.013
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47–e47. https://doi.org/10.1093/nar/gkv007
Rogers NH, Witczak CA, Hirshman MF, Goodyear LJ, Greenberg AS (2009) Estradiol stimulates Akt, AMP-activated protein kinase (AMPK) and TBC1D1/4, but not glucose uptake in rat soleus. Biochem Biophys Res Commun 382:646–650. https://doi.org/10.1016/j.bbrc.2009.02.154
Roy D, Cai Q, Felty Q, Narayan S (2007) Estrogen-induced generation of reactive oxygen and nitrogen species, gene damage, and estrogen-dependent cancers. J Toxicol Enviro Health B 10:235–257. https://doi.org/10.1080/15287390600974924
RStudio Team (2020) RStudio: integrated development environment for R. R Studio, PBC, Boston
Rushing BR, Selim MI (2019) Aflatoxin B1: a review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem Toxicol 124:81–100. https://doi.org/10.1016/j.fct.2018.11.047
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Song H (2004) Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J Gastroenterol 10:509. https://doi.org/10.3748/wjg.v10.i4.509
Sprague Dawley. https://www.albany.edu/mcnaylab/sd.html [accessed 31 Jan 2023].
Storvik M, Huuskonen P, Kyllönen T, Lehtonen S, El-Nezami H, Auriola S et al (2011) Aflatoxin B1—a potential endocrine disruptor—up-regulates CYP19A1 in JEG-3 cells. Toxicol Lett 202:161–167. https://doi.org/10.1016/j.toxlet.2011.01.028
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Tang Y, Li M, Wang J, Pan Y, Wu F-X (2015) CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 127:67–72. https://doi.org/10.1016/j.biosystems.2014.11.005
Teo AKC, Oh HK, Ali RB, Li BFL (2001) The modified human DNA repair enzyme O6-methylguanine-DNA methyltransferase is a negative regulator of estrogen receptor-mediated transcription upon alkylation DNA damage. Mol Cell Biol 21:7105–7114. https://doi.org/10.1128/mcb.21.20.7105-7114.2001
The GIMP Development Team. 2019. GIMP.
US Government. 2002. Electronic code of federal regulations (e-CFR), 7 CFR Part 996—minimum quality and handling standards for domestic and imported peanuts marketed in the United States. 996.11—negative aflatoxin content. https://www.law.cornell.edu/cfr/text/7/996.11 [accessed 23 Jan 2022].
Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E (2021) gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput 17:6281–6291. https://doi.org/10.1021/acs.jctc.1c00645
Wan L, O’Brien P (2014) Molecular mechanism of 17alpha-ethinylestradiol cytotoxicity in isolated rat hepatocytes. Can J Physiol Pharmacol 92:21–26. https://doi.org/10.1139/cjpp-2013-0267
Xing D, Oparil S, Yu H, Gong K, Feng W, Black J et al (2012) Estrogen modulates NFkappaB signaling by enhancing Ikappa B alpha levels and blocking p65 binding at the promoters of inflammatory genes via estrogen receptor-beta: S. Kovats, ed. PLoS ONE 7:e36890. https://doi.org/10.1371/journal.pone.0036890
Zamir-Nasta T, Razi M, Shapour H, Malekinejad H (2017) Roles of p21, p53, cyclin D1, CDK-4, estrogen receptor alpha in aflatoxin B1-induced cytotoxicity in testicular tissue of mice. Environ Toxicol 33:385–395. https://doi.org/10.1002/tox.22524
Zhan T, Rindtorff N, Boutros M (2017) Wnt signaling in cancer. Oncogene 36:1461–1473. https://doi.org/10.1038/onc.2016.304
Acknowledgements
We thank Mr. Adam Harris for providing technical support. JUV was the recipient of an ERASMUS Student Mobility Traineeship. GH acknowledges support from US NIH U54MD010706, U01DA045300, and QUB start-up funds.
Funding
This study was supported by the Bualuang ASEAN Chair Professor Fund (CTE).
Author information
Authors and Affiliations
Contributions
JUV, CP, WDAS, RP and GH conceived the experiment(s). JUV and CP conducted the experiments. JUV, CP, WDAS, RP and GH analyzed the results. JUV and GH drafted the manuscript. JUV, CP, WDAS, ESH, APN, CTE, OC, RP and GH edited the manuscript. OC, RP, and GH provided supervision.
Corresponding authors
Ethics declarations
Competing interest
G.H. is a founder of Altomics Datamation Ltd. and a member of its scientific advisory board.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Verga, J.U., Padovano, C., da Silveira, W.A. et al. A Systems Biology Approach Reveals the Endocrine Disrupting Potential of Aflatoxin B1. Expo Health 16, 321–340 (2024). https://doi.org/10.1007/s12403-023-00557-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12403-023-00557-w