Abstract
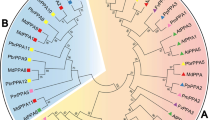
OVATE family proteins (OFPs) are the plant-specific transcription factors, and have significant functions in regulating plant growth, development and resistance. The OFP genes have been investigated in several plants, but they still lack a systematic analysis of OFP genes in Chinese pear and some other five Rosaceae genomes. Here, 28 PbrOFPs were identified within Chinese pear and compared them with those of other five Rosaceae genomes. Evolutionary tree revealed that all OFP genes from six Rosaceae genomes were divided into eight groups. Seventeen conserved microsynteny regions were detected in Chinese pear genome, suggested that these PbrOFP genes might be considered to have originated from the large-scale duplication events., indicating these PbrOFP genes might contain specialized regulatory mechanisms in these tissues, such as flower, ovary and fruit. Remarkably, two PbrOFP genes (Pbr010426.1 and Pbr010427.1) were up-regulated under Venturia nashicola treatment, and five PbrOFP genes were up-regulated under PEG treatment, suggesting that these genes might play crucial roles in defence to environmental stresses. Our data presented a systematic analysis and might aid in the selection of appropriate PbrOFPs for further functional studies in Chinese pear, especially in response to the mechanism of biotic and abiotic stresses.






Similar content being viewed by others
References
Cai B, Yang X, Tuskan GA, Cheng Z-M (2011) MicroSyn: a user-friendly tool for detection of microsynteny in a gene family. BMC Bioinform 12:79
Cao Y, Han Y, Li D, Lin Y, Cai Y, Fips J (2016) MYB transcription factors in Chinese pear (Pyrus bretschneideri Rehd.): genome-wide identification, classification, and expression profiling during fruit development. Front Plant Sci 7:577
Cao Y, Li X, Jiang L (2019a) Integrative analysis of the core fruit lignification toolbox in pear reveals targets for fruit quality bioengineering. Biomolecules 9:504
Cao Y et al (2019b) Integrative analysis reveals evolutionary patterns and potential functions of SWEET transporters in Euphorbiaceae. Int J Biol Macromol 139:1–11
Cao Y, Xu X, Jiang L (2020) Integrative analysis of the RNA interference toolbox in two Salicaceae willow species, and their roles in stress response in poplar (Populus trichocarpa Torr. & Gray). Int J Biol Macromol 162:1127–1139. https://doi.org/10.1016/j.ijbiomac.2020.06.235
Chagné D et al (2014) The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS One 9:e92644
Daccord N et al (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49:1099–1106
Fan C, Guo G, Yan H, Qiu Z, Liu Q, Zeng B (2018) Characterization of Brassinazole resistant (BZR) gene family and stress-induced expression in Eucalyptus grandis. Physiol Mol Biol Plants 24:821–831
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucl Acids Res 39:W29–W37
Ghorbani R, Zakipour Z, Alemzadeh A, Razi H (2020) Genome-wide analysis of AP2/ERF transcription factors family in Brassica napus. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-020-00832-z
Hackbusch J, Richter K, Mller J, Salamini F, Uhrig JF (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci 102:4908–4912
Huang Z, Van Houten J, Gonzalez G, Xiao H, van der Knaap E (2013) Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol Genet Genom 288:111–129
Jones P et al (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kavas M, Kızıldoğan AK, Balık HI (2019) Gene expression analysis of bud burst process in European hazelnut (Corylus avellana L.) using RNA-Seq. Physiol Mol Biol Plants 25:13–29
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357
Letunic I, Doerks T, Bork P (2011) SMART 7: recent updates to the protein domain annotation resource. Nucl Acids Res 40:D302–D305
Li E, Wang S, Liu Y, Chen JG, Douglas CJ (2011) Ovate Family Protein4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J 67:328–341
Li H, Dong Q, Zhao Q, Ran K (2019) Genome-wide identification, expression profiling, and protein-protein interaction properties of ovate family proteins in apple. Tree Genet Genom 15:45
Liu Y, Douglas CJ (2015) A role for Ovate family protein1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in Arabidopsis thaliana. Plant Signal Behav 10:e1033126
Liu J, Van Eck J, Cong B, Tanksley SD (2002) A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci 99:13302–13306
Liu J et al (2015) Banana Ovate family protein MaOFP1 and MADS-box protein MuMADS1 antagonistically regulated banana fruit ripening. PLoS ONE 10:e0123870
Ma Y, Yang C, He Y, Tian Z, Li J, Sunkar R (2017) Rice OVATE family protein 6 regulates plant development and confers resistance to drought and cold stresses. J Exp Bot 68:4885–4898
Pagnussat GC, Yu H-J, Sundaresan V (2007) Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19:3578–3592
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11:1650
Punta M et al (2011) The Pfam protein families database. Nucl Acids Res 40:D290–D301
Schmitz AJ, Begcy K, Sarath G, Walia H (2015) Rice Ovate Family Protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci 241:177–188
Shafiei-Koij F, Ravichandran S, Barthet VrJ, Rodrigue N, Mirlohi A, Majidi MM, Cloutier S (2020) Evolution of Carthamus species revealed through sequence analyses of the fad2 gene family. Physiol Mol Biol Plants 26:419–432
Shulaev V et al (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43:109–116
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Verde I et al (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45:487–494
Wang S, Chang Y, Guo J, Chen JG (2007) Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J 50:858–872
Wang Y-K, Chang W-C, Liu P-F, Hsiao M-K, Lin C-T, Lin S-M, Pan R-L (2010) Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair. Plant Mol Biol 74:453–466
Wang S, Chang Y, Guo J, Zeng Q, Ellis BE, Chen J-G (2011) Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS ONE 6:e23896
Wang S, Chang Y, Ellis B (2016) Overview of Ovate family proteins, a novel class of plant-specific growth regulators. Front Plant Sci 7:417
Wu J et al (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Yu H, Jiang W, Liu Q, Zhang H, Piao M, Chen Z, Bian M (2015) Expression pattern and subcellular localization of the ovate protein family in rice. PLoS ONE 10:e0118966
Zhang Q et al (2018) The genetic architecture of floral traits in the woody plant Prunus mume. Nat Commun 9:1–12
Funding
The funding was provided by the Shanxi Province Science Foundation for Youths (Grant No. 201801D221290).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethical approval
This article did not include any experiments with human participants or animals performed by all the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ding, B., Hu, C., Feng, X. et al. Systematic analysis of the OFP genes in six Rosaceae genomes and their roles in stress response in Chinese pear (Pyrus bretschneideri). Physiol Mol Biol Plants 26, 2085–2094 (2020). https://doi.org/10.1007/s12298-020-00866-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-020-00866-3