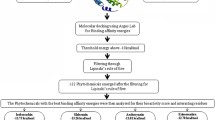
Abstract
Prostate cancer is the World's second most frequent malignancy, with the fifth-highest male mortality rate. In advanced prostate cancer patients, point mutations such as T877A and W741L are prevalent, imparting treatment resistance and hence promoting cancer development. The emergence of drug resistance in prostate cancer necessitates the development of suitable ligands to allow for stronger interactions with the receptors, which can inhibit cancer progression. The present study focuses on flavonoids produced by plants, which may act as inhibitors of point mutations like T877A and W741L in prostate cancer. This research was conducted using an in-silico method where the compound Glabranin and its derivatives were virtually screened to identify potential drugs for combating such point mutations. Thirty-five Molecular Dockings were performed to find the ligand-receptor complexes with the lowest binding energy. Moreover, employing a variety of tools, ligands were evaluated for drug-likeness and toxicity, indicating a promising drug candidate. Based on the results of Molecular Docking, Drug-likeness, and ADMET testing, eight structures were subjected to a 100 ns Molecular Dynamics simulation. A QSAR analysis was also performed based on the simulation findings. In this study, it was revealed that GlaMod2 phytocompound was effective against T877A and W741L mutations in prostate cancer. It was observed that the phytocompound was stable and had potential properties for the development of a novel drug to combat prostate cancer and drug resistance This phytocompound may therefore be effective in the development of prostate cancer inhibitors for patients with mutant androgen receptors.
Graphical Abstract





Similar content being viewed by others
Availability of Data and Materials
All data generated or analyzed during this study are included in this manuscript and its online resource file.
References
Batiha GES, Beshbishy AM, El-Mleeh A, Abdel-Daim MM, Devkota HP. Traditional uses bioactive chemical constituents, and pharmacological and toxicological activities of Glycyrrhiza glabra L Fabaceae. Biomolecules. 2020;10(3):352. https://doi.org/10.3390/biom10030352.
Kreyling WG, Semmler M, Erbe F, Mayer P, Takenaka S, Schulz H, et al. Translocation of ultrafine insoluble iridium particles from lung epithelium to extrapulmonary organs is size dependent but very low. J Toxicol Environ Health A. 2002;65(20):1513–30. https://doi.org/10.1080/00984100290071649.
El-Saber Batiha G, Magdy Beshbishy A, El-Mleeh A, Abdel-Daim MM, Prasad Devkota H. Traditional uses, bioactive chemical constituents, and pharmacological and toxicological activities of Glycyrrhiza glabra L. (Fabaceae). Biomolecules. 2020;10(3):352. https://doi.org/10.3390/biom10030352.
Wang L, Yang R, Yuan B, Liu Y, Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm Sin B. 2005;5(4):310–5. https://doi.org/10.1016/j.apsb.2015.05.005.
Varshney IP, Jain DC, Srivastava HC. Study of Saponins from Glycyrrhiza glabra Root. Int J Crude Drug Res. 1983;21(4):169–72. https://doi.org/10.3109/13880208309070637.
Shinde DB, Koratkar SS, Sharma N, Shitole AA. Antioxidant activity and antiproliferative action of methanolic extract of liquorice (Glycyrrhiza glabra) in HEPG2 cell line. Int J Pharm Pharm Sci. 2016. https://doi.org/10.22159/ijpps.2016v8i9.11954
Govind P, Sharma M. Ethnomedicinal plants for prevention and treatment of tumors. Int J Green Pharm. 2009;3(1):2. https://doi.org/10.4103/0973-8258.49367.
Pilcher H. Liquorice may tackle SARS. Nature. 2003. https://doi.org/10.1038/news030609-16.
Young S-M, Bansal P, Vella ET, Finelli A, Levitt C, Loblaw A. Systematic review of clinical features of suspected prostate cancer in primary care. Can Fam Physician. 2015;61(1):e26-35.
Kaler J, Hussain A, Haque A, Naveed H, Patel SA. Comprehensive review of pharmaceutical and surgical interventions of prostate cancer. Cureus. 2020;12(11):e11617. https://doi.org/10.7759/cureus.11617.
Chen J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harbor Perspectives in medicine 2016;6(3), a026104. https://doi.org/10.1101/cshperspect.a026104.
Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2(4):466–74. https://doi.org/10.1177/1947601911408889.
Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinogenesis. 2011;10:20. https://doi.org/10.4103/1477-3163.83937.
Rathkopf DE, Smith MR, Ryan CJ, Berry WR, Shore ND, Liu G, Higano CS, Alumkal JJ, Hauke R, Tutrone RF, Saleh M, Chow Maneval E, Thomas S, Ricci DS, Yu MK, de Boer CJ, Trinh A, Kheoh T, Bandekar R, Scher HI, Antonarakis ES. Androgen receptor mutations in patients with castration-resistant prostate cancer treated with apalutamide. Annals Oncol Off J European Soci Med Oncol. 2017;28(9):2264–71. https://doi.org/10.1093/annonc/mdx283.
Sakkiah S, Kusko R, Pan B, Guo W, Ge W, Tong W, Hong H. Structural changes due to antagonist binding in ligand binding pocket of androgen receptor elucidated through molecular dynamics simulations. Front Pharmacol. 2018. https://doi.org/10.3389/fphar.2018.00492.
Nadiminty N, Gao AC. Mechanisms of persistent activation of the androgen receptor in CRPC: Recent advances and future perspectives. World J Urol. 2012; 30, 287–295.
Terakawa T, Miyake H, Kumano M, Sakai I, Fujisawa M. The antiandrogen bicalutamide activates the androgen receptor (AR) with a mutation in codon 741 through the mitogen activated protein kinase (MARK) pathway in human prostate cancer PC3 cells. Oncol Rep. 2010;24(5):1395–9. https://doi.org/10.3892/or_00000998.
Singh AN, Baruah MM, Sharma N. Structure-Based docking studies towards exploring the potential anti-androgen activity of selected phytochemicals against Prostate Cancer. Sci Rep. 1955; 7(1). https://doi.org/10.1038/s41598-017-02023-5.
Tian S, Wang J, Li Y, Li D, Xu L, Hou T. The application of in silico drug-likeness predictions in pharmaceutical research. Adv Drug Deliv Rev. 2015;86:2–10. https://doi.org/10.1016/j.addr.2015.01.009.
Salsbury FR. Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Curr Opin Pharmacol. 2010;10(6):738–44. https://doi.org/10.1016/j.coph.2010.09.016.
Hansch C, Verma RP. Overcoming tumor drug resistance with C2-modified 10-deacetyl-7-propionyl cephalomannines: a QSAR study. Mol Pharm. 2009;6(3):849–60. https://doi.org/10.1021/mp800138w.
Bohl CE, Miller DD, Chen J, Bell CE, Dalton JT. Structural basis for accommodation of nonsteroidal ligands in the androgen receptor. J Biol Chem. 2005;280(45):37747–54. https://doi.org/10.1074/jbc.M507464200.
Bohl CE, Miller DD, Chen J, Bell CE, Dalton JT. Structural basis for antagonism and resistance of bicalutamide in prostate cancer. Proc Natl Acad Sci. 2005;102(17):6201–6. https://doi.org/10.1073/pnas.0500381102.
Matias PM. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J Biol Chem. 2000;275(34):26164–71. https://doi.org/10.1074/jbc.m004571200.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. https://doi.org/10.1002/jcc.20084.
National Center for Biotechnology Information. PubChem Compound Summary for CID 124049, Glabranin. Retrieved March 17, 2023 from https://pubchem.ncbi.nlm.nih.gov/compound/Glabranin.
National Center for Biotechnology Information. PubChem Compound Summary for CID 91649, Hydroxyflutamide. Retrieved March 17, 2023 from https://pubchem.ncbi.nlm.nih.gov/compound/Hydroxyflutamide.
National Center for Biotechnology Information. PubChem Compound Summary for CID 56069, (R)-Bicalutamide. Retrieved March 17, 2023 from https://pubchem.ncbi.nlm.nih.gov/compound/R_-Bicalutamide.
National Center for Biotechnology Information. PubChem Compound Summary for CID 2375, Bicalutamide. Retrieved March 17, 2023 from https://pubchem.ncbi.nlm.nih.gov/compound/Bicalutamide.
National Center for Biotechnology Information. PubChem Compound Summary for CID 3397, Flutamide. Retrieved March 17, 2023 from https://pubchem.ncbi.nlm.nih.gov/compound/Flutamide.
ChemAxon - Software Solutions and Services for Chemistry & Biology. Chemaxon.com. 2021. https://chemaxon.com/. Accessed 15 Dec 2021.
Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179(3):269–78. https://doi.org/10.1016/j.jsb.2011.09.006.
Biswas SS, Browne RB, Borah VV, Roy JD. In Silico approach for phytocompound-based drug designing to fight efflux pump-mediated multidrug-resistant mycobacterium tuberculosis. Appl Biochem Biotechnol. 2021;193(6):1757–79. https://doi.org/10.1007/s12010-021-03557-1.
Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. In: Chemical biology, 2015. pp. 243–250. Humana Press, New York, NY.
Ramezani A, Zakeri A, Mard-Soltani M, Mohammadian A, Hashemi ZS, Mohammadpour H, Jahangiri A, Khalili S, Rasaee MJ. Structure based screening for inhibitory therapeutics of CTLA-4 unveiled new insights about biology of ACTH. Int J Peptide Res Therapeut. 2019. https://doi.org/10.1007/s10989-019-09891-7.
Souza PF, Lopes FE, Amaral JL, Freitas CD, Oliveira JT. A molecular docking study revealed that synthetic peptides induced conformational changes in the structure of SARS-CoV-2 spike glycoprotein, disrupting the interaction with human ACE2 receptor. Int J Biol Macromol. 2020;164:66–76.
Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–86.
Molinspiration Cheminformatics. Choice (Middletown). 2006. 43(11):43-6538-43-6538. https://www.molinspiration.com/. Accessed 15 Dec 2021.
Drug Likeness Tool (DruLiTo 1). Gov. in. 2021. http://www.niper.gov.in/pi_dev_tools/ DruLiToWeb/DruLiTo_index.html. Accessed 15 Dec 2021.
Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–86. https://doi.org/10.1021/ci200227u.
Bekker H, Berendsen HJC, Dijkstra EJ, Achterop S, Vondrumen R, Vanderspoel D, Sijbers A, Keegstra H, Renardus MKR. Gromacs-a parallel computer for molecular-dynamics simulations. In: 4th International Conference on Computational Physics (PC 92). 1993; pp. 252–256. World Scientific Publishing.
Kumari R, Kumar R. Open-source drug discovery consortium, Lynn A. g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–62. https://doi.org/10.1021/ci500020m.
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, et al. 3rd, calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–97. https://doi.org/10.1021/ar000033j.
Davies M, Nowotka M, Papadatos G, Dedman N, Gaulton A, Atkinson F, et al. ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015;43(W1):W612–20. https://doi.org/10.1093/nar/gkv352.
Gerçek Z, Ceyhan D, Erçağ E. Synthesis and molecular docking study of novel COVID-19 inhibitors. Turk J Chem. 2021;45(3):704. https://doi.org/10.3906/kim-2012-55.
Kuntal. EasyQSAR: A beginners tool for QSAR in Drug Designing (Free software for drug designing and QSAR). Blogspot.com. 2021. http://easyqsar.blogspot.com/2009/09/easyqsar-beginners-tool-for-qsar-in.html. Accessed 16 Dec 2021.
Banu BH, Rajitha G, Bharathi K. An approach to computer aided drug design of some bioactive cinnamoyl hydrazones, in silico and docking studies as possible COX-2 selective inhibitors. J Biotechnol Biomater. 2016. https://doi.org/10.4172/2155-952x.1000240.
Leeson PD, Davis AM. Time-related differences in the physical property profiles of oral drugs. J Med Chem. 2004;47(25):6338–48. https://doi.org/10.1021/jm049717d.
Muchmore SW, Edmunds JJ, Stewart KD, Hajduk PJ. Cheminformatic tools for medicinal chemists. J Med Chem. 2010;53:4830–41.
Selvam P, De S, Paira P, Kumar SA, Moorthy A, Ghosh A, Jenifer SK. In vitro studies on the selective cytotoxic effect of luminescent Ru (II)-p-cymene complexes of imidazo-pyridine and imidazo quinoline ligands. Dalton Trans. 2022;51(45):17263–76.
De S, Kumar RS, Gauthaman A, Kumar SA, Paira P, Moorthy A, Banerjee S. Luminescent ruthenium(II)-para-cymene complexes of aryl substituted imidazo-1,10-phenanthroline as anticancer agents and the effect of remote substituents on cytotoxic activities. Inorganica Chimica Acta. 2021;515:120066. https://doi.org/10.1016/j.ica.2020.120066.
Pandey B, Grover A, Sharma P. Molecular dynamics simulations revealed structural differences among WRKY domain-DNA interaction in barley (Hordeum vulgare). BMC Genom. 2018. https://doi.org/10.1186/s12864-018-4506-3.
Browne RB, Goswami N, Borah P, Roy JD. Computational approaches for evaluation of isobavachin as potential inhibitor against t877a and w741l mutations in prostate cancer. J Biomol Struct Dyn. 2022; https://doi.org/10.1080/07391102.2022.2032353.
Fujita T, Iwasa J, Hansch C. A new substituent constant, π, derived from partition coefficients. J Am Chem Soc. 1964;86(23):5175–80.
Abdel-Ilah L, Veljović E, Gurbeta L, Badnjević A. Applications of QSAR study in drug design. Int J Eng Res Technol (IJERT). 2017; 6(06).
Fujita K, Nonomura N. Role of androgen receptor in prostate cancer: a review. World J Mens Health. 2019;37(3):288–95. https://doi.org/10.5534/wjmh.180040.
Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20(13):3001–15. https://doi.org/10.1200/jco.2002.10.018.
Acknowledgements
We acknowledge the Bioinformatics Infrastructure Facility, C.V.Sc., Khanapara, Guwahati, sponsored by the Department of Biotechnology, Ministry of Science and Technology, Govt. of India for providing facilities to conduct molecular dynamics studies in a high-performance Dell Precision Tesla 5600 workstation with 12 CPUs and 1 NVIDIA Tesla C2075 graphics processing unit (GPU). We would also like to thank the Vice Chancellor, Assam Don Bosco University for providing the research facilities. The authors appreciate the continuous encouragement and advice from the Director, School of Life Science, Assam Don Bosco University.
Funding
The authors declare that no funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
The manuscript was written through the contributions of all authors. All authors have approved the final version of the manuscript. R. B. B. has designed and compiled the Manuscript. J. D. R. has conceived and supervised the research. N. G. has guided the computational work and also in performing the Molecular Dynamics Simulation studies. P. B. who is the Coordinator, BIF, and Head of the Department of Animal Biotechnology, College of Veterinary Science has guided and allowed to perform molecular dynamics simulation studies in a high-performance workstation in his lab. All these above-mentioned authors contributed equally to the successful completion of the research work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Browne, R.B., Goswami, N., Borah, P. et al. Study of Glabranin as an Inhibitor Against Prostate Cancer: Molecular Docking, Molecular Dynamics Simulation, MM-PBSA Calculation and QSAR Prediction. Ind J Clin Biochem (2023). https://doi.org/10.1007/s12291-023-01134-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12291-023-01134-3