Abstract
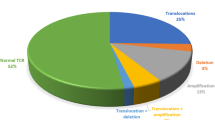
T cell acute lymphoblastic leukaemia (T-ALL) is a genetically heterogeneous and aggressive form of malignancy. Although a number of recurrent fusion genes are reported in T-ALL, their involvement in disease stratification and therapeutic intervention is still controversial. Considering the prognostic value of STIL-TAL1 fusion and tyrosine kinase inhibitor (TKI) based therapeutic potential of NUP214-ABL1, the present study aimed to investigate their frequency and clinical correlation in pediatric T-ALL cases. Our cohort consisted of 48 T-ALL pediatric cases (age ≤ 12 years) with a median age of 6 years and male to female ratio of 20.5:1. The median TLC of the study group was noted to be 220,000/ cu mm with a range from 26,810/cu mm to 785,430/cu mm. By MLPA and RT-PCR analysis we observed that 11/48 cases (23%) showed STIL-TAL1 fusion and 4/48 cases (8.3%) had NUP214-ABL1 fusion gene. Both of the fusion genes did not show any significant correlation with any of the clinico-hematological or treatment outcome parameters. However, upon analysis of copy number variations (CNVs) with other clinically relevant genes, we found significant correlation between LEF1 (p = 0.024) and PTEN (p = 0.049) gene deletions with STIL/TAL1 fusion in T-ALL patients. NUP214-ABL1 fusion gene did not reveal any association with either CNVs or with survival. Although limited with the small cohort size and follow up, our study supports the similar frequency of these fusions as compared to other Asian and Western studies and also highlights utility of MLPA technique as a good diagnostic modality to screen for both STIL-TAL1 and NUP214-ABL1 fusions in a single assay with additional data on secondary copy number changes.

Similar content being viewed by others
References
Pui C-H, Robison LL, Look AT (2008) Acute lymphoblastic leukaemia. Lancet (London, England). Vol. 371. pp 1030–1043. http://linkinghub.elsevier.com/retrieve/pii/S0140673608604572. Accessed 3 Jan 2008
Hebert J, Cayuela JM, Berkeley J, Sigaux F (1994) Candidate tumor-suppressor genes MTS1 (p16INK4A) and MTS2 (p15INK4B) display frequent homozygous deletions in primary cells from T- but not from B-cell lineage acute lymphoblastic leukemias. Blood. Vol. 84, pp 4038–4044. http://www.ncbi.nlm.nih.gov/pubmed/7994022. Accessed 17 Jun 2018
Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al (2002) Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. Vol. 1. pp 75–87. http://www.ncbi.nlm.nih.gov/pubmed/12086890. Accessed 3 Jun 2018
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. (2018) Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. Vol. 306. pp 269–71. http://www.sciencemag.org/cgi. doi/https://doi.org/10.1126/science.1102160. Accesed 13 Jun 2018
Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. (2009) Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncolited. Vol. 10. pp 147–56. http://linkinghub.elsevier.com/retrieve/pii/S1470204508703140. Accessed 17 Jun 2018
Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J et al (2012) Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366:1079–1089
Apperley JF (2015) Chronic myeloid leukaemia. Lancet. Vol. 385. pp 1447–59. https://linkinghub.elsevier.com/retrieve/pii/S0140673613621200. Accessed 5 Nov 2019
Inaba H, Greaves M, Mullighan CG (2013) Acute lymphoblastic leukaemia. Lancet 381:1943–1955
Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, et al. (2001) Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. Vol. 344. pp 1038–42. http://www.ncbi.nlm.nih.gov/pubmed/11287973. Accessed 5 Nov 2019
Site-specific Recombination of the tal-1 gene is a common occurrence in human T cell Leukemia – PubMed. https://pubmed.ncbi.nlm.nih.gov/2209547/. Accessed 13 Jul 2020
De Braekeleer E, Douet-Guilbert N, Rowe D, Bown N, Morel F, Berthou C, et al. (2011) ABL1 fusion genes in hematological malignancies: a review. Eur J Haematol, Wiley. Vol. 86. pp 361–71. http://doi.wiley.com/https://doi.org/10.1111/j.1600-0609.2011.01586.x. Accessed 13 Jul 2020
Graux C, Stevens-Kroef M, Lafage M, Dastugue N, Harrison CJ, Mugneret F, et al. (2020) Heterogeneous patterns of amplification of the NUP214-ABL1 fusion gene in T-cell acute lymphoblastic leukemia. Leukemia . Nature Publishing Group. Vol. 23. pp 125–33. www.nature.com/leu. Accessed 13 Jul 2020
Bhatia P, Totadri S, Singh M, Sharma P, Trehan A, Bansal D, et al. (2020) PEST domain NOTCH mutations confer a poor relapse free survival in pediatric T-ALL: data from a tertiary care centre in India. Blood Cells, Mol Dis. Academic Press. 2020. Vol. 82. https://pubmed.ncbi.nlm.nih.gov/32179411/. Accessed 13 Jul 2020
Van Dongen JJM, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, et al. (1999) Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 concerted action: investigation of minimal residual disease in acute leukemia. Leukemia. Nature Publishing Group. p. 1901–1928. https://pubmed.ncbi.nlm.nih.gov/10602411/. Accessed 13 Jul 2020
Thakral D, Kaur G, Gupta R, Benard-Slagter A, Savola S, Kumar I, et al. (2019) Rapid identification of key copy number alterations in B- and T-cell acute lymphoblastic leukemia by digital multiplex ligation-dependent probe amplification. Front Oncol. Frontiers Media S.A. Vol. 9. https://pubmed.ncbi.nlm.nih.gov/31572674/. Accessed 13 Jul 2020
Chen B, Jiang L, Zhong ML, Li JF, Li BS, Peng LJ, et al. (2017) Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. Nat Acad Sci. Vol. 115. pp 373–378. https://pubmed.ncbi.nlm.nih.gov/29279377/. Accessed 13 Jul 2020
D’Angiò M, Valsecchi MG, Testi AM, Conter V, Nunes V, Parasole R, et al. (2013) Clinical features and outcome of SIL/TAL1-positive t-cell acute lymphoblastic leukemia in children and adolescents: a 10-year experience of the AIEOP group. Haematologica. Ferrata Storti Foundation. p. e10–3. Available from: www.haematologica.org. Accessed 13 Jul 2020
Tesio M, Trinquand A, Ballerini P, Lhermitte L, Petit A, Ifrah N et al (2017) Age-related clinical and biological features of PTEN abnormalities in T-cell acute lymphoblastic leukaemia. Leukemia 31:2594–2600. https://doi.org/10.1038/leu.2017.15
Furness CL, Mansur MB, Weston VJ, Ermini L, van Delft FW, Jenkinson S, et al. (2018) The subclonal complexity of STIL-TAL1+ T-cell acute lymphoblastic leukaemia. Leukemia. Nature Publishing Group. Vol. 32. pp 1984–1993. https://pubmed.ncbi.nlm.nih.gov/29556024/. Accessed 13 Jul 2020
Kimura S, Seki M, Yoshida K, Ueno H, Shiraishi Y, Chiba K et al (2016) TAL1 super enhancer aberration and stil-TAL1 fusion in pediatric T cell acute lymphoblastic leukemia. Blood Am Soc Hematol 128:1734–1734
Wang D, Zhu G, Wang N, Zhou X, Yang Y, Zhou S, et al. (2013) SIL-TAL1 rearrangement is related with poor outcome: a study from a Chinese institution. PLoS One. PLoS One. Vol. 8. https://pubmed.ncbi.nlm.nih.gov/24040098/. Accessed 13 Jul 2020
Mansur MB, Emerenciano M, Brewer L, Sant’Ana M, Mendonça N, Thuler LCS, et al. (2020) SIL-TAL1 fusion gene negative impact in T-cell acute lymphoblastic leukemia outcome. Leuk Lymphoma. Leuk Lymphoma. Vol. 50. pp 1318–25. https://pubmed.ncbi.nlm.nih.gov/19562638/. Accessed 13 Jul 2020
Noronha EP, Codeço Marques LV, Andrade FG, Santos Thuler LC, Terra-Granado E, Pombo-De-Oliveira MS (2020) The profile of immunophenotype and genotype aberrations in subsets of pediatric T-cell acute lymphoblastic leukemia. Front Oncol. Frontiers Media S.A. Vol. 9. https://pubmed.ncbi.nlm.nih.gov/31338319/. Accessed 13 Jul 2020
Duployez N, Grzych G, Ducourneau B, Fuentes MA, Grardel N, Boyer T, et al. (2020) NUP214-ABL1 fusion defines a rare subtype of B-cell precursor acute lymphoblastic leukemia that could benefit from tyrosine kinase inhibitors. Haematologica. Ferrata Storti Foundation; 2016. p e133–4. https://pubmed.ncbi.nlm.nih.gov/26681761/. Accessed 13 Jul 2020
Funding
Special Research Grant, PGIMER.
Author information
Authors and Affiliations
Contributions
PB conceptualised the study; NP, MS, PS performed the experiments; PB, NP, MS analysed the data; PB, NP performed statistical analysis; AT, SN, DB provided patient sample and clinical data; PB, MS wrote and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None.
Consent to Participate
Yes.
Consent for Publication
Yes.
Availability of Data and Material
Not applicable.
Ethical Approval
Yes.
Code Availability
Not Applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patra, N., Singh, M., Sharma, P. et al. Clinico-Hematological Profile and Copy Number Abnormalities in a Cohort of STIL-TAL1 and NUP214-ABL1 Positive Pediatric T-Cell Acute Lymphoblastic Leukemia. Indian J Hematol Blood Transfus 37, 555–562 (2021). https://doi.org/10.1007/s12288-020-01394-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12288-020-01394-6