Abstract
Background
Next-generation sequencing (NGS) has enabled comprehensive genomic profiling to identify gene alterations that play important roles in cancer biology. However, the clinical significance of these genomic alterations in triple-negative breast cancer (TNBC) patients has not yet been fully elucidated. The aim of this study was to clarify the clinical significance of genomic profiling data, including copy number alterations (CNA) and tumor mutation burden (TMB), in TNBC patients.
Methods
A total of 47 patients with Stage I–III TNBC with genomic profiling of 435 known cancer genes by NGS were enrolled in this study. Disease-free survival (DFS) and overall survival (OS) were evaluated for their association to gene profiling data.
Results
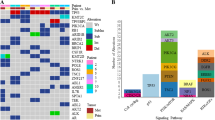
CNA-high patients showed significantly worse DFS and OS than CNA-low patients (p = 0.0009, p = 0.0041, respectively). TMB was not associated with DFS or OS in TNBC patients. Patients with TP53 alterations showed a tendency of worse DFS (p = 0.0953) and significantly worse OS (p = 0.0338) compared with patients without TP53 alterations. Multivariable analysis including CNA and other clinicopathological parameters revealed that CNA was an independent prognostic factor for DFS (p = 0.0104) and OS (p = 0.0306). Finally, multivariable analysis also revealed the combination of CNA-high and TP53 alterations is an independent prognostic factor for DFS (p = 0.0005) and OS (p = 0.0023).
Conclusions
We revealed that CNA, but not TMB, is significantly associated with DFS and OS in TNBC patients. The combination of CNA-high and TP53 alterations may be a promising biomarker that can inform beyond standard clinicopathologic factors to identify a subgroup of TNBC patients with significantly worse prognosis.



Similar content being viewed by others
Data availability
The data will be shared on reasonable request to TW.
References
Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–8.
Pal SK, Childs BH, Pegram M. Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat. 2011;125:627–36.
Irvin WJ Jr, Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44:2799–805.
Zagami P, Carey LA. Triple negative breast cancer: Pitfalls and progress. NPJ Breast Cancer. 2022;8:95.
Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7.
Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60.
Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. 2014; 3.
Shlien A, Raine K, Fuligni F, Arnold R, Nik-Zainal S, Dronov S, et al. Direct transcriptional consequences of somatic mutation in Breast Cancer. Cell Rep. 2016;16:2032–46.
Bertucci F, Chaffanet M, Birnbaum D. An ICGC major achievement in breast cancer: a comprehensive catalog of mutations and mutational signatures. Chin Clin Oncol. 2017;6:4.
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X, et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell. 2019;35:428-40.e5.
Nagahashi M, Shimada Y, Ichikawa H, Kameyama H, Takabe K, Okuda S, et al. Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci. 2019;110:6–15.
Nagahashi M, Wakai T, Shimada Y, Ichikawa H, Kameyama H, Kobayashi T, et al. Genomic landscape of colorectal cancer in Japan: clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med. 2016;8:136.
Yuza K, Nagahashi M, Watanabe S, Takabe K, Wakai T. Hypermutation and microsatellite instability in gastrointestinal cancers. Oncotarget. 2017;8:112103–15.
Ichikawa H, Wakai T, Nagahashi M, Shimada Y, Hanyu T, Kano Y, et al. Pathogenic germline BRCA1/2 mutations and familial predisposition to gastric cancer. JCO PO. 2018; 2:PO.18.00097.
Nagahashi M, Sato S, Yuza K, Shimada Y, Ichikawa H, Watanabe S, et al. Common driver mutations and smoking history affect tumor mutation burden in lung adenocarcinoma. J Surg Res. 2018;230:181–5.
Tsuchida J, Rothman J, McDonald KA, Nagahashi M, Takabe K, Wakai T. Clinical target sequencing for precision medicine of breast cancer. Int J Clin Oncol. 2019;24:131–40.
Nagahashi M, Ling Y, Hayashida T, Kitagawa Y, Futamura M, Yoshida K, et al. Actionable gene alterations in an Asian population with triple-negative breast cancer. JCO PO. 2018; 2:PO.17.00211.
Edge SB, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC cancer staging manual. 7th ed. New York: Springer; 2010.
Kurosumi M. Immunohistochemical assessment of hormone receptor status using a new scoring system (J-Score) in breast cancer. Breast Cancer. 2007;14:189–93.
Allison KH, Hammond MEH, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL, et al. Estrogen and progesterone receptor testing in Breast Cancer: ASCO/CAP guideline update. J Clin Oncol. 2020;38:1346–66.
Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, et al. Human epidermal growth factor receptor 2 testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol. 2018;36:2105–22.
Nagahashi M, Shimada Y, Ichikawa H, Nakagawa S, Sato N, Kaneko K, et al. Formalin-fixed paraffin-embedded sample conditions for deep next generation sequencing. J Surg Res. 2017;220:125–32.
Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12: e1004873.
Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9.
Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54.
Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular Breast Cancer. Cell. 2015;163:506–19.
Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol. 2018;15:139–50.
Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ, et al. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47:490–9.
Zhang Y, Martens JW, Yu JX, Jiang J, Sieuwerts AM, Smid M, et al. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69:3795–801.
Nedeljković M, Tanić N, Dramićanin T, Milovanović Z, Šušnjar S, Milinković V, et al. Importance of copy number alterations of FGFR1 and C-MYC genes in triple negative Breast cancer. J Med Biochem. 2019;38:63–70.
Suelmann BBM, Rademaker A, van Dooijeweert C, van der Wall E, van Diest PJ, Moelans CB. Genomic copy number alterations as biomarkers for triple negative pregnancy-associated breast cancer. Cell Oncol (Dordr). 2022;45:591–600.
El Ansari FZ, Jouali F, Fekkak R, Bakkach J, Ghailani Nourouti N, Barakat A, et al. BRCA1/2 variants and copy number alterations status in non familial triple negative breast cancer and high grade serous ovarian cancer. Hered Cancer Clin Pract. 2022;20:29.
Zhang L, Feizi N, Chi C, Hu P. Association Analysis of Somatic Copy Number Alteration Burden With Breast Cancer Survival. Front Genet. 2018;9:421.
Stover DG, Parsons HA, Ha G, Freeman SS, Barry WT, Guo H, et al. Association of cell-free DNA tumor fraction and somatic copy number alterations with survival in metastatic triple-negative Breast Cancer. J Clin Oncol. 2018;36:543–53.
Li Z, Zhang X, Hou C, Zhou Y, Chen J, Cai H, et al. Comprehensive identification and characterization of somatic copy number alterations in triple-negative breast cancer. Int J Oncol. 2020;56:522–30.
Kobayashi S. Basal-like subtype of breast cancer: a review of its unique characteristics and their clinical significance. Breast Cancer. 2008;15:153–8.
Duffy MJ, Synnott NC, Crown J. Mutant p53 in breast cancer: potential as a therapeutic target and biomarker. Breast Cancer Res Treat. 2018;170:213–9.
Sukumar J, Gast K, Quiroga D, Lustberg M, Williams N. Triple-negative breast cancer: promising prognostic biomarkers currently in development. Expert Rev Anticancer Ther. 2021;21:135–48.
Koçak A, Heselmeyer-Haddad K, Lischka A, Hirsch D, Fiedler D, Hu Y, et al. High levels of chromosomal copy number alterations and TP53 mutations correlate with poor outcome in Younger Breast Cancer Patients. Am J Pathol. 2020;190:1643–56.
Gao C, Li H, Liu C, Xu X, Zhuang J, Zhou C, et al. Tumor mutation burden and immune invasion characteristics in triple negative Breast Cancer: genome high-throughput data analysis. Front Immunol. 2021;12: 650491.
Tan Q, Yin S, Zhou D, Chi Y, Man X, Li H. Potential predictive and prognostic value of biomarkers related to immune checkpoint inhibitor therapy of triple-negative Breast Cancer. Front Oncol. 2022;12: 779786.
Ke L, Li S, Cui H. The prognostic role of tumor mutation burden on survival of breast cancer: a systematic review and meta-analysis. BMC Cancer. 2022;22:1185.
Acknowledgements
We thank ClearScience (http://www.clearscience.net/) for English language editing.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research Grant Number 22H03140 and 21K19522 for MN, and research funding from Denka for WT and SO.
Author information
Authors and Affiliations
Contributions
Conception and design: All authors; Administrative support: MN, YS, HI, YM, SO, TW; Collection and assembly of data: MN, YL, CT, TH, YK, MF, TK, SN, HY, TY, KK, CK, NS, JT, KM, MN; Data analysis and interpretation: MN, YL, SL, YM, KT, SO, TW; Writing the first draft of manuscript: MN, SL, YM, KT, SO, TW; Review and editing: All authors; Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Conflict of interest
M Nagahashi received honoraria from Chugai, AstraZeneca, Eli Lilly, Pfizer, Novartis, Taiho, Daiichi Sankyo, Esai, Kyowa-Kirin, and Denka. T Hayashida received research funding and honoraria from Eisai, Chugai, Eli Lilly, Pfizer, Kyowa-Kirin, Daiichi-Sankyo, Fixstars and Taiho. Y Kitagawa received grants and personal fees from ASAHI KASEI PHARMA CORPORATION, grants, personal fees and other from ONO PHARMACEUTICAL CO., LTD., grants and personal fees from Otsuka Pharmaceutical Factory, Inc., grants and personal fees from Nippon Covidien Inc., grants, personal fees and other from TAIHO PHARMACEUTICAL CO., LTD, grants, personal fees and other from CHUGAI PHARMACEUTICAL CO., LTD., grants and personal fees from KAKEN PHARMACEUTICAL CO.,LTD., personal fees from AstraZeneca K.K., personal fees from Ethicon Inc., personal fees from Olympus Corporation, personal fees from SHIONOGI & CO., LTD., personal fees and other from Bristol-Myers Squibb K.K., personal fees from MSD K.K., personal fees from Smith & Nephew KK, personal fees from ASKA Pharmaceutical Co., Ltd., personal fees from MIYARISAN PHARMACEUTICAL CO. LTD., personal fees from Toray Industries, Inc., personal fees from DAIICHI SANKYO COMPANY, LIMITED, personal fees from Chugai Foundation for Innovative Drug Discovery Science, personal fees from Nippon Kayaku Co., Ltd., grants from Yakult Honsha Co. Ltd., grants from Otsuka Pharmaceutical Co., Ltd., grants from TSUMURA & CO., grants from Sumitomo Pharma Co., Ltd., grants from EA Pharma Co., Ltd., grants from Eisai Co., Ltd., grants from Kyowa Kirin Co.,Ltd., grants from MEDICON INC., grants from Takeda Pharmaceutical Co., Ltd., grants from TEIJIN PHARMA LIMITED., outside the submitted work. H Yamauchi received research funding from AstraZeneca, and EIKEN Kagaku. N Sato received honorarium from Chugai Pharmaceutical, Kyowa Kirin Co. Ltd., Taiho Pharmaceutical, Eli Lilly, Nippon Kayaku Co., Daiichi Sankyo, and Celltrion Healthcare Japan. S Lyle is a paid consultant for KEW Inc. Y Miyoshi received research funding and honoraria from Esai, Chugai, AstraZeneca, Eli Lilly, Pfizer, MSD, Kyowa-Kirin, Daiichi-Sankyo, and Taiho. T Wakai, and S Okuda received research funding from Denka.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Nagahashi, M., Ling, Y., Toshikawa, C. et al. Copy number alteration is an independent prognostic biomarker in triple-negative breast cancer patients. Breast Cancer 30, 584–595 (2023). https://doi.org/10.1007/s12282-023-01449-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-023-01449-2



