Abstract
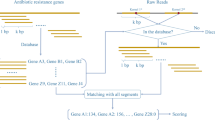
Whole genome and metagenome sequencing are powerful approaches that enable comprehensive cataloging and profiling of antibiotic resistance genes at scales ranging from a single clinical isolate to ecosystems. Recent studies deal with genomic and metagenomic data sets at larger scales; therefore, designing computational workflows that provide high efficiency and accuracy is becoming more important. In this review, we summarize the computational workflows used in the research field of antibiotic resistome based on genome or metagenome sequencing. We introduce workflows, software tools, and data resources that have been successfully employed in this rapidly developing field. The workflow described in this review can be used to list the known antibiotic resistance genes from genomes and metagenomes, quantitatively profile them, and investigate the epidemiological and evolutionary contexts behind their emergence and transmission. We also discuss how novel antibiotic resistance genes can be discovered and how the association between the resistome and mobilome can be explored.
Similar content being viewed by others
References
Alcock, B.P., Raphenya, A.R., Lau, T.T.Y., Tsang, K.K., Bouchard, M., Edalatmand, A., Huynh, W., Nguyen, A.L.V., Cheng, A.A., Liu, S., et al. 2019. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525.
Angiuoli, S.V. and Salzberg, S.L. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27, 334–342.
Antonopoulos, D.A., Assaf, R., Aziz, R.K., Brettin, T., Bun, C., Conrad, N., Davis, J.J., Dietrich, E.M., Disz, T., Gerdes, S., et al. 2019. PATRIC as a unique resource for studying antimicrobial resistance. Brief. Bioinform. 20, 1094–1102.
Arango-Argoty, G., Garner, E., Pruden, A., Heath, L.S., Vikesland, P., and Zhang, L. 2018. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6, 23.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A.A., Dvorkin, M., Kulikov, A.S., Lesin, V.M., Nikolenko, S.I., Pham, S., Prjibelski, A.D., et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477.
Bayliss, S.C., Thorpe, H.A., Coyle, N.M., Sheppard, S.K., and Feil, E.J. 2019. PIRATE: A fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. GigaScience 8, giz119.
Beghini, F., McIver, L.J., Blanco-Míguez, A., Dubois, L., Asnicar, F., Maharjan, S., Mailyan, A., Thomas, A.M., Manghi, P., Valles-Colomer, M., et al. 2020. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bio-Bakery 3. bioRxiv. doi: https://doi.org/10.1101/2020.11.19.388223
Berglund, F., Böhm, M.E., Martinsson, A., Ebmeyer, S., Österlund, T., Johnning, A., Larsson, D.G.J., and Kristiansson, E. 2020. Comprehensive screening of genomic and metagenomic data reveals a large diversity of tetracycline resistance genes. Microb. Genom. 6, mgen000455.
Berglund, F., Österlund, T., Boulund, F., Marathe, N.P., Larsson, D.G.J., and Kristiansson, E. 2019. Identification and reconstruction of novel antibiotic resistance genes from metagenomes. Microbiome 7, 52.
Böhm, M.E., Razavi, M., Marathe, N.P., Flach, C.F., and Larsson, D.G.J. 2020. Discovery of a novel integron-borne aminoglycoside resistance gene present in clinical pathogens by screening environmental bacterial communities. Microbiome 8, 41.
Bortolaia, V., Kaas, R.S., Ruppe, E., Roberts, M.C., Schwarz, S., Cattoir, V., Philippon, A., Allesoe, R.L., Rebelo, A.R., Florensa, A.F., et al. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500.
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C.H., Xie, D., Suchard, M.A., Rambaut, A., and Drummond, A.J. 2014. BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537.
Bradley, P., Gordon, N.C., Walker, T.M., Dunn, L., Heys, S., Huang, B., Earle, S., Pankhurst, L.J., Anson, L., de Cesare, M., et al. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun. 6, 10063.
Břinda, K., Callendrello, A., Ma, K.C., MacFadden, D.R., Charalampous, T., Lee, R.S., Cowley, L., Wadsworth, C.B., Grad, Y.H., Kucherov, G., et al. 2020. Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing. Nat. Microbiol. 5, 455–464.
Brito, I.L., Yilmaz, S., Huang, K., Xu, L., Jupiter, S.D., Jenkins, A.P., Naisilisili, W., Tamminen, M., Smillie, C.S., Wortman, J.R., et al. 2016. Mobile genes in the human microbiome are structured from global to individual scales. Nature 535, 435–439.
Brown, C.T., Moritz, D., O’Brien, M.P., Reidl, F., Reiter, T., and Sullivan, B.D. 2020. Exploring neighborhoods in large metagenome assembly graphs using spacegraphcats reveals hidden sequence diversity. Genome Biol. 21, 164.
Buchfink, B., Xie, C., and Huson, D.H. 2015. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60.
Campbell, T.P., Sun, X., Patel, V.H., Sanz, C., Morgan, D., and Dantas, G. 2020. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 14, 1584–1599.
Chaumeil, P.A., Mussig, A.J., Hugenholtz, P., and Parks, D.H. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927.
Chen, S., Zhou, Y., Chen, Y., and Gu, J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890.
Cohen, K.A., Manson, A.L., Desjardins, C.A., Abeel, T., and Earl, A.M. 2019. Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges. Genome Med. 11, 45.
Croucher, N.J., Page, A.J., Connor, T.R., Delaney, A.J., Keane, J.A., Bentley, S.D., Parkhill, J., and Harris, S.R. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15.
Darling, A.E., Mau, B., and Perna, N.T. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5, e11147.
Didelot, X., Croucher, N.J., Bentley, S.D., Harris, S.R., and Wilson, D.J. 2018. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 46, e134.
Didelot, X., Fraser, C., Gardy, J., and Colijn, C. 2017. Genomic infectious disease epidemiology in partially sampled and ongoing outbreaks. Mol. Biol. Evol. 34, 997–1007.
Doyle, R.M., O’Sullivan, D.M., Aller, S.D., Bruchmann, S., Clark, T., Coello Pelegrin, A., Cormican, M., Diez Benavente, E., Ellington, M.J., McGrath, E., et al. 2020. Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: an inter-laboratory study. Microb. Genom. 6, e000335.
Ellington, M.J., Ekelund, O., Aarestrup, F.M., Canton, R., Doumith, M., Giske, C., Grundman, H., Hasman, H., Holden, M.T.G., Hopkins, K.L., et al. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 23, 2–22.
Feldgarden, M., Brover, V., Haft, D.H., Prasad, A.B., Slotta, D.J., Tolstoy, I., Tyson, G.H., Zhao, S., Hsu, C.H., McDermott, P.F., et al. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63, e00483.
Gasparrini, A.J., Wang, B., Sun, X., Kennedy, E.A., Hernandez-Leyva, A., Ndao, I.M., Tarr, P.I., Warner, B.B., and Dantas, G. 2019. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 4, 2285–2297.
Ghaly, T.M., Geoghegan, J.L., Alroy, J., and Gillings, M.R. 2019. High diversity and rapid spatial turnover of integron gene cassettes in soil. Environ. Microbiol. 21, 1567–1574.
Gupta, S.K., Padmanabhan, B.R., Diene, S.M., Lopez-Rojas, R., Kempf, M., Landraud, L., and Rolain, J.M. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220.
Joensen, K.G., Tetzschner, A.M.M., Iguchi, A., Aarestrup, F.M., and Scheutz, F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426.
Jolley, K.A., Bray, J.E., and Maiden, M.C.J. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124.
Kent, A.G., Vill, A.C., Shi, Q., Satlin, M.J., and Brito, I.L. 2020. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 11, 4379.
Kim, J., Greenberg, D.E., Pifer, R., Jiang, S., Xiao, G., Shelburne, S.A., Koh, A., Xie, Y., and Zhan, X. 2020. VAMPr: VAriant Mapping and Prediction of antibiotic resistance via explainable features and machine learning. PLoS Comput. Biol. 16, e1007511.
Kim, D., Song, L., Breitwieser, F.P., and Salzberg, S.L. 2016. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 26, 1721–1729.
Kolmogorov, M., Bickhart, D.M., Behsaz, B., Gurevich, A., Rayko, M., Shin, S.B., Kuhn, K., Yuan, J., Polevikov, E., Smith, T.P.L., et al. 2020. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110.
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P.A. 2019. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546.
Kothari, A., Wu, Y.W., Chandonia, J.M., Charrier, M., Rajeev, L., Rocha, A.M., Joyner, D.C., Hazen, T.C., Singer, S.W., and Mukhopadhyay, A. 2019. Large circular plasmids from groundwater plasmidomes span multiple incompatibility groups and are enriched in multimetal resistance genes. mBio 10, e02899–18.
Lee, K., Kim, D.W., Lee, D.H., Kim, Y.S., Bu, J.H., Cha, J.H., Thawng, C.N., Hwang, E.M., Seong, H.J., Sul, W.J., et al. 2020. Mobile resistome of human gut and pathogen drives anthropogenic bloom of antibiotic resistance. Microbiome 8, 2.
Lees, J.A., Harris, S.R., Tonkin-Hill, G., Gladstone, R.A., Lo, S.W., Weiser, J.N., Corander, J., Bentley, S.D., and Croucher, N.J. 2019. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 29, 304–316.
Li, D., Liu, C.M., Luo, R., Sadakane, K., and Lam, T.W. 2015. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676.
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., Tai, C., Deng, Z., and Ou, H.Y. 2019. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665.
MacFadden, D.R., Coburn, B., Břinda, K., Corbeil, A., Daneman, N., Fisman, D., Lee, R.S., Lipsitch, M., McGeer, A., Melano, R.G., et al. 2020. Using genetic distance from archived samples for the prediction of antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 64, e02417–19.
Mahé, P. and Tournoud, M. 2018. Predicting bacterial resistance from whole-genome sequences using k-mers and stability selection. BMC Bioinformatics 19, 383.
Marçais, G., Delcher, A.L., Phillippy, A.M., Coston, R., Salzberg, S.L., and Zimin, A. 2018. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 14, e1005944.
Marttinen, P., Hanage, W.P., Croucher, N.J., Connor, T.R., Harris, S.R., Bentley, S.D., and Corander, J. 2012. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 40, e6.
Meier-Kolthoff, J.P. and Göker, M. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10, 2182.
Minh, B.Q., Schmidt, H.A., Chernomor, O., Schrempf, D., Woodhams, M.D., von Haeseler, A., and Lanfear, R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534.
Munck, C., Sheth, R.U., Freedberg, D.E., and Wang, H.H. 2020. Recording mobile DNA in the gut microbiota using an Escherichia coli CRISPR-Cas spacer acquisition platform. Nat. Commun. 11, 95.
Nguyen, M., Long, S.W., McDermott, P.F., Olsen, R.J., Olson, R., Stevens, R.L., Tyson, G.H., Zhao, S., and Davis, J.J. 2019. Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J. Clin. Microbiol. 57, e01260.
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P.A. 2017. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 27, 824–834.
Olekhnovich, E.I., Vasilyev, A.T., Ulyantsev, V.I., Kostryukova, E.S., and Tyakht, A.V. 2018. MetaCherchant: analyzing genomic context of antibiotic resistance genes in gut microbiota. Bioinformatics 34, 434–444.
Oliveira, P.H., Touchon, M., Cury, J., and Rocha, E.P.C. 2017. The chromosomal organization of horizontal gene transfer in bacteria. Nat. Commun. 8, 841.
Ondov, B.D., Treangen, T.J., Melsted, P., Mallonee, A.B., Bergman, N.H., Koren, S., and Phillippy, A.M. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132.
Page, A.J., Cummins, C.A., Hunt, M., Wong, V.K., Reuter, S., Holden, M.T., Fookes, M., Falush, D., Keane, J.A., and Parkhill, J. 2015. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693.
Page, A.J., Taylor, B., and Keane, J.A. 2016. Multilocus sequence typing by blast from de novo assemblies against PubMLST. J. Open Source Softw. 1, 118.
Parks, D.H., Chuvochina, M., Waite, D.W., Rinke, C., Skarshewski, A., Chaumeil, P.A., and Hugenholtz, P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004.
Perez-Sepulveda, B.M., Heavens, D., Pulford, C.V., Predeus, A.V., Low, R., Webster, H., Schudoma, C., Rowe, W., Lipscombe, J., Watkins, C., et al. 2020. An accessible, efficient and global approach for the large-scale sequencing of bacterial genomes. bioRxiv 200840. doi: https://doi.org/10.1101/2020.07.22.200840.
Price, M.N., Dehal, P.S., and Arkin, A.P. 2010. FastTree 2 — approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490.
Robertson, J. and Nash, J.H.E. 2018. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4, e000206.
Ruppé, E., Ghozlane, A., Tap, J., Pons, N., Alvarez, A.S., Maziers, N., Cuesta, T., Hernando-Amado, S., Clares, I., Martínez, J.L., et al. 2019. Prediction of the intestinal resistome by a three-dimensional structure-based method. Nat. Microbiol. 4, 112–123.
Sayers, E.W., Beck, J., Brister, J.R., Bolton, E.E., Canese, K., Comeau, D.C., Funk, K., Ketter, A., Kim, S., Kimchi, A., et al. 2020. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 48, D9–D16.
Schwengers, O., Barth, P., Falgenhauer, L., Hain, T., Chakraborty, T., and Goesmann, A. 2020. Platon: identification and characterization of bacterial plasmid contigs in short-read draft assemblies exploiting protein sequence-based replicon distribution scores. Microb. Genom. 6, mgen000398.
Segerman, B. 2020. The most frequently used sequencing technologies and assembly methods in different time segments of the bacterial surveillance and RefSeq genome databases. Front. Cell. Infect. Microbiol. 10, 527102.
Sheppard, A.E., Stoesser, N., Wilson, D.J., Sebra, R., Kasarskis, A., Anson, L.W., Giess, A., Pankhurst, L.J., Vaughan, A., Grim, C.J., et al. 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob. Agents Chemother. 60, 3767–3778.
Shi, W., Sun, Q., Fan, G., Hideaki, S., Moriya, O., Itoh, T., Zhou, Y., Cai, M., Kim, S.G., Lee, J.S., et al. 2020. gcType: a high-quality type strain genome database for microbial phylogenetic and functional research. Nucleic Acids Res. 49, D694–D705.
Smillie, C.S., Smith, M.B., Friedman, J., Cordero, O.X., David, L.A., and Alm, E.J. 2011. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244.
Spencer, S.J., Tamminen, M.V., Preheim, S.P., Guo, M.T., Briggs, A.W., Brito, I.L., Weitz, D.A., Pitkänen, L.K., Vigneault, F., Virta, M.P.J., et al. 2016. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 10, 427–436.
Steinegger, M., Mirdita, M., and Söding, J. 2019. Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat. Methods 16, 603–606.
Steinegger, M. and Söding, J. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028.
Treangen, T.J., Ondov, B.D., Koren, S., and Phillippy, A.M. 2014. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524.
Van Camp, P.J., Haslam, D.B., and Porollo, A. 2020. Prediction of antimicrobial resistance in Gram-negative bacteria from whole-genome sequencing data. Front. Microbiol. 11, 1013.
Wallace, J.C., Port, J.A., Smith, M.N., and Faustman, E.M. 2017. FARME DB: a functional antibiotic resistance element database. Database 2017, baw165.
Wang, R., van Dorp, L., Shaw, L.P., Bradley, P., Wang, Q., Wang, X., Jin, L., Zhang, Q., Liu, Y., Rieux, A., et al. 2018. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9, 1179.
Wick, R.R., Heinz, E., Holt, K.E., and Wyres, K.L. 2018. Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 56, e00197–18.
Wick, R.R. and Holt, K.E. 2020. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Res. 8, 2138.
Wood, D.E., Lu, J., and Langmead, B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 257.
Xie, Z. and Tang, H. 2017. ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33, 3340–3347.
Yin, X., Jiang, X.T., Chai, B., Li, L., Yang, Y., Cole, J.R., Tiedje, J.M., and Zhang, T. 2018. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 34, 2263–2270.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617.
Yoshida, C.E., Kruczkiewicz, P., Laing, C.R., Lingohr, E.J., Gannon, V.P.J., Nash, J.H.E., and Taboada, E.N. 2016. The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE 11, e0147101.
Yuan, Q.B., Huang, Y.M., Wu, W.B., Zuo, P., Hu, N., Zhou, Y.Z., and Alvarez, P.J.J. 2019. Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ. Int. 131, 104986.
Zankari, E., Allesøe, R., Joensen, K.G., Cavaco, L.M., Lund, O., and Aarestrup, F.M. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768.
Zhang, A.N., Li, L.G., Ma, L., Gillings, M.R., Tiedje, J.M., and Zhang, T. 2018. Conserved phylogenetic distribution and limited antibiotic resistance of class 1 integrons revealed by assessing the bacterial genome and plasmid collection. Microbiome 6, 130.
Zhou, Z., Alikhan, N.F., Mohamed, K., Fan, Y., Agama Study Group, and Achtman, M. 2019. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 30, 138–152.
Acknowledgments
This work was supported by the Korea Ministry of Environment (MOE) as “the Environmental Health Action Program (2016001350004)” and by the “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ015130032020)” Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of Interest
The authors declare that they have no conflict of interest
Rights and permissions
About this article
Cite this article
Lee, K., Kim, DW. & Cha, CJ. Overview of bioinformatic methods for analysis of antibiotic resistome from genome and metagenome data. J Microbiol. 59, 270–280 (2021). https://doi.org/10.1007/s12275-021-0652-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-021-0652-4