Abstract
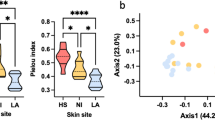
Acne vulgaris, commonly known as acne, is the most common skin disorder and a multifactorial disease of the sebaceous gland. Although the pathophysiology of acne is still unclear, bacterial and fungal factors are known to be involved in. This study aimed to investigate whether the microbiomes and mycobiomes of acne patients are distinct from those of healthy subjects and to identify the structural signatures of microbiomes related to acne vulgaris. A total of 33 Korean female subjects were recruited (Acne group, n = 17; Healthy group, n = 16), and microbiome samples were collected swabbing the forehead and right cheek. To characterize the fungal and bacterial communities, 16S rRNA V4-V5 and ITS1 region, respectively, were sequenced and analysed using Qiime2. There were no significant differences in alpha and beta diversities of microbiomes between the Acne and Healthy groups. In comparison with the ratio of Cutibacterium to Staphylococcus, the acne patients had higher abundance of Staphylococcus compared to Cutibacterium than the healthy individuals. In network analysis with the dominant microorganism amplicon sequence variants (ASV) (Cutibacterium, Staphylococcus, Malassezia globosa, and Malassezia restricta) Cutibacterium acnes was identified to have hostile interactions with Staphylococcus and Malassezia globosa. Accordingly, this results suggest an insight into the differences in the skin microbiome and mycobiome between acne patients and healthy controls and provide possible microorganism candidates that modulate the microbiomes associated to acne vulgaris.
Similar content being viewed by others
References
Akaza, N., Akamatsu, H., Numata, S., Yamada, S., Yagami, A., Nakata, S., and Matsunaga, K. 2016. Microorganisms inhabiting follicular contents of facial acne are not only Propionibacterium but also Malassezia spp. J. Dermatol. 43, 906–911.
Alexeyev, O.A., Lundskog, B., Ganceviciene, R., Palmer, R.H., McDowell, A., Patrick, S., Zouboulis, C., and Golovleva, I. 2012. Pattern of tissue invasion by Propionibacterium acnes in acne vulgaris. J. Dermatol. Sci. 67, 63–66.
Amir, A., McDonald, D., Navas-Molina, J.A., Kopylova, E., Morton, J.T., Zech Xu, Z., Kightley, E.P., Thompson, L.R., Hyde, E.R., Gonzalez, A., et al. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191–16.
Barnard, E., Shi, B., Kang, D., Craft, N., and Li, H. 2016. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 6, 39491.
Belkaid, Y. and Segre, J.A. 2014. Dialogue between skin microbiota and immunity. Science 346, 954–959.
Bhambri, S., Del Rosso, J.Q., and Bhambri, A. 2009. Pathogenesis of acne vulgaris: recent advances. J. Drugs Dermatol. 8, 615–618.
Bokulich, N.A., Subramanian, S., Faith, J.J., Gevers, D., Gordon, J.I., Knight, R., Mills, D.A., and Caporaso, J.G. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59.
Byrd, A.L., Belkaid, Y., and Segre, J.A. 2018. The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155.
Caporaso, J.G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F.D., Costello, E.K., Fierer, N., Peña, A.G., Goodrich, J.K., Gordon, J.I., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Chen, Y.E. and Tsao, H. 2013. The skin microbiome: current perspectives and future challenges. J. Am. Acad. Dermatol. 69, 143–155.
Dagnelie, M.A., Montassier, E., Khammari, A., Mounier, C., Corvec, S., and Dréno, B. 2019. Inflammatory skin is associated with changes in the skin microbiota composition on the back of severe acne patients. Exp. Dermatol. 28, 961–967.
Dréno, B., Martin, R., Moyal, D., Henley, J.B., Khammari, A., and Seité, S. 2017. Skin microbiome and acne vulgaris: Staphylococcus, a new actor in acne. Exp. Dermatol. 26, 798–803.
Fitz-Gibbon, S., Tomida, S., Chiu, B.H., Nguyen, L., Du, C., Liu, M., Elashoff, D., Erfe, M.C., Loncaric, A., Kim, J., et al. 2013. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol. 133, 2152–2160.
Grice, E.A., Kong, H.H., Renaud, G., Young, A.C., NISC Comparative Sequencing Program, Bouffard, G.G., Blakesley, R.W., Wolfsberg, T.G., Turner, M.L., and Segre, J.A. 2008. A diversity profile of the human skin microbiota. Genome Res. 18, 1043–1050.
Kelhälä, H.L., Aho, V.T.E., Fyhrquist, N., Pereira, P.A.B., Kubin, M.E., Paulin, L., Palatsi, R., Auvinen, P., Tasanen, K., and Lauerma, A. 2018. Isotretinoin and lymecycline treatments modify the skin microbiota in acne. Exp. Dermatol. 27, 30–36.
Kurtz, Z.D., Müller, C.L., Miraldi, E.R., Littman, D.R., Blaser, M.J., and Bonneau, R.A. 2015. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11, e1004226.
Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 17, 10–12.
Nilsson, R.H., Larsson, K.H., Taylor, A.F.S., Bengtsson-Palme, J., Jeppesen, T.S., Schigel, D., Kennedy, P., Picard, K., Glöckner, F.O., Tedersoo, L., et al. 2019. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264.
O’Neill, A.M. and Gallo, R.L. 2018. Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome 6, 177.
Park, T., Kim, H.J., Myeong, N.R., Lee, H.G., Kwack, I., Lee, J., Kim, B.J., Sul, W.J., and An, S. 2017. Collapse of human scalp microbiome network in dandruff and seborrhoeic dermatitis. Exp. Dermatol. 26, 835–838.
Rivers, A.R., Weber, K.C., Gardner, T.G., Liu, S., and Armstrong, S.D. 2018. ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Res. 7, 1418.
Sanford, J.A. and Gallo, R.L. 2013. Functions of the skin microbiota in health and disease. Semin. Immunol. 25, 370–377.
Xu, H. and Li, H. 2019. Acne, the skin microbiome, and antibiotic treatment. Am. J. Clin. Dermatol. 20, 335–344.
Acknowledgments
This research was supported by a grant from the Chung-Ang University Graduate Research Scholarship in 2019, and by the AmorePacific Co. R&D Center, and the National Research Foundation of Korea (NRF) grants funded by the South Korean Government (2019R1A4A1024764).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest We have no conflicts of interest to report.
Ethical Statement This project was reviewed by the Institutional Review Board (IRB) of the Korea National Institute for Bioethics Policy (IRB no. IECK(1)-IRB-18K041326), and written consent was provided by all subjects.
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kim, J., Park, T., Kim, HJ. et al. Inferences in microbial structural signatures of acne microbiome and mycobiome. J Microbiol. 59, 369–375 (2021). https://doi.org/10.1007/s12275-021-0647-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-021-0647-1