Abstract
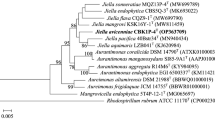
A Gram-negative, aerobic, short-rod-shaped, motile (with a terminal flagellum), non-spore-forming bacterium, designated strain 85T, was isolated from a surface-sterilized bark of Sonneratia caseolaris collected from Qinzhou in Guangxi, China and was analyzed using a polyphasic approach to determine its taxonomic position. Strain 85T grew optimally in the presence of 1–2% (w/v) NaCl at 30°C and pH 6.0–7.0. Phylogenetic analysis based on 16S rRNA gene sequence suggested that strain 85T belonged to the genus Fulvimarina and shared the highest 16S rRNA gene sequence similarity with Fulvimarina pelagi HTCC2506T (96.16%). The cell-wall peptidoglycan contained meso-diaminopimelic acid and ubiquinone Q-10 was the predominant respiratory lipoquinone. The polar lipids comprised diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, an unidentified amino lipid, three unidentified phospholipids and six unidentified lipids. The major fatty acid was C18:1ω7c. The DNA G+C content of strain 85T was 65.4 mol%, and the average nucleotide identity and estimated DDH values between strain 85T and the type strain of Fulvimarina pelagi HTCC2506T were 77.3% and 21.7%, respectively. Based on the phylogenetic, phenotypic, and chemotaxonomic analyses, strain 85T should be considered as a novel species of the genus Fulvimarina with the proposed name Fulvimarina endophytica sp. nov., and its type strain is 85T (= KCTC 62717T = CGMCC 1.13665T).
Similar content being viewed by others
References
Cappuccino, J.G. and Sherman, N. 2002. Microbiology: a laboratory manual, 6th ed. Benjamin Cummings Pearson Education, San Francisco, USA.
Cho, J.C. and Giovannoni, S.J. 2003. Fulvimarina pelagi gen. nov., sp. nov., a marine bacterium that forms a deep evolutionary lineage of descent in the order “Rhizobiales”. Int. J. Syst. Evol. Microbiol. 53, 1853–1859.
Collins, M.D., Pirouz, T., Goodfellow, M., and Minnikin, D.E. 1977. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230.
Denner, E.B., Smith, G.W., Busse, H.J., Schumann, P., Narzt, T., Polson, S.W., Lubitz, W., and Richardson, L.L. 2003. Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean scleractinian corals. Int. J. Syst. Evol. Microbiol. 53, 1115–1122.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Fitch, W.M. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20, 406–416.
Gonzalez, C., Gutierrez, C., and Ramirez, C. 1978. Halobacterium vallismortis sp. nov., an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can. J. Microbiol. 24, 710–715.
Guo, L., Tuo, L., Habden, X., Zhang, Y., Liu, J., Jiang, Z., Liu, S., Dilbar, T., and Sun, C. 2015. Allosalinactinospora lopnorensis gen. nov., sp. nov., a new member of the family Nocardiopsaceae isolated from soil. Int. J. Syst. Evol. Microbiol. 65, 206–213.
Kelly, K.L. 1964. Inter-society color council-national bureau of standards color name charts illustrated with centroid colors. US Government Printing Office, Washington, DC, USA.
Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J. Mol. Evol. 16, 111–120.
Li, W.J., Xu, P., Schumann, P., Zhang, Y.Q., Pukall, R., Xu, L.H., Stackebrandt, E., and Jiang, C.L. 2007. Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int. J. Syst. Evol. Microbiol. 57, 1424–1428.
Liang, J., Liu, J., and Zhang, X.H. 2015. Jiella aquimaris gen. nov., sp. nov., isolated from offshore surface seawater. Int. J. Syst. Evol. Microbiol. 65, 1127–1132.
Magee, C.M., Rodeheaver, G., Edgerton, M.T., and Edlich, R.F. 1975. A more reliable Gram staining technic for diagnosis of surgical infections. Am. J. Surg. 130, 341–346.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Parks, D.H., Imelfort, M., Skennerton, C.T., Hugenholtz, P., and Tyson, G.W. 2015. Check M: assessing the quality of microbial genomes recovered from isolateds, single cells, and metagenomes. Genome Res. 25, 1043–1055.
Qin, S., Wang, H.B., Chen, H.H., Zhang, Y.Q., Jiang, C.L., Xu, L.H., and Li, W.J. 2008. Glycomyces endophyticus sp. nov., an endophytic actinomycete isolated from the root of Carex baccans Nees. Int. J. Syst. Evol. Microbiol. 58, 2525–2528.
Rathsack, K., Reitner, J., Stackebrandt, E., and Tindall, B.J. 2011. Reclassification of Aurantimonas altamirensis (Jurado et al. 2006), Aurantimonas ureilytica (Weon et al. 2007) and Aurantimonas frigidaquae (Kim et al. 2008) as members of a new genus, Aureimonas gen.nov., as Aureimonas altamirensis gen. nov., comb. nov., Aureimonas ureilytica comb. nov. and Aureimonas frigidaquae comb. nov., and emended descriptions of the genera Aurantimonas and Fulvimarina. Int. J. Syst. Evol. Microbiol. 61, 2722–2728.
Ren, F., Zhang, L., Song, L., Xu, S., Xi, L., Huang, L., Huang, Y., and Dai, X. 2014. Fulvimarina manganoxydans sp. nov., isolated from a deep-sea hydrothermal plume in the south-west Indian ocean. Int. J. Syst. Evol. Microbiol. 64, 2920–2925.
Richter, M. and Rosselló-Móra, R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131.
Rivas, R., Sánchez-Márquez, S., Mateos, P.F., Martínez-Molina, E., and Velázquez, E. 2005. Martelella mediterranea gen. nov., sp. nov., a novel alpha-proteobacterium isolated from a subterranean saline lake. Int. Syst. Evol. Microbiol. 55, 955–959.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc. Newark, DE, USA.
Schleifer, K.H. and Kandler, O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477.
Shirling, E.B. and Gottlieb, D. 1966. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882.
Xu, P., Li, W.J., Tang, S.K., Zhang, Y.Q., Chen, G.Z., Chen, H.H., Xu, L.H., and Jiang, C.L. 2005. Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int. J. Syst. Evol. Microbiol. 55, 1149–1153.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (NSFC, Grant no. 81603079), Science and Technology Foundation of Guizhou Province (No. Qian Ke He Jichu[2019]1347) and Youth Science and technology personnal growth project of Guizhou Provincial Education Department (No. Qian Jiao He KY Zi[2016]200).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Tuo, L., Yan, XR. Fulvimarina endophytica sp. nov., a novel endophytic bacterium isolated from bark of Sonneratia caseolaris. J Microbiol. 57, 655–660 (2019). https://doi.org/10.1007/s12275-019-8627-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-019-8627-4