Abstract
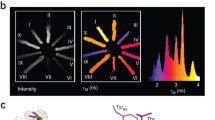
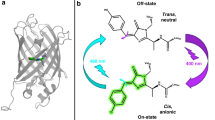
The Keima family comprises large Stokes shifts fluorescent proteins, which are useful for dual-color fluorescence crosscorrelation spectroscopy and multicolor imaging. dKeima570 belongs to the Keima family. It has a unique chromophore sequence composed of CYG with an emission peak at 570 nm, but its molecular properties are unclear. We report the spectral analysis of dKeima570 and its crystal structure at 2.0 Å resolution. The dKeima570 chromophore is mainly in the protonation state in the entire pH range. The pH-induced non-fluorescence state was observed below pH 4.0. The crystal structure of the dKeima570 chromophore has a cis conformation at pH 6.5. The chromophore is surrounded by a unique hydrogen bonding network containing a water bridge between Glu212 and Arg194. The analysis of the dimeric interface of dKeima570 revealed the key residues that maintain the oligomerization of Keima family. Structural comparisons of dKeima570 and mKeima provided insights into the unique large Stokes shifts characteristics of the Keima family.
Similar content being viewed by others
References
Adams, P.D., Afonine, P.V., Bunkoczi, G., Chen, V.B., Davis, I.W., Echols, N., Headd, J.J., Hung, L.W., Kapral, G.J., Grosse-Kunstleve, R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221.
Bae, J.E., Kim, I.J., and Nam, K.H. 2017. Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221. Biochem. Biophys. Res. Commun. 493, 562–567.
Bae, J.E., Kim, I.J., and Nam, K.H. 2018. Spectroscopic analysis of the Cu2+-induced fluorescence quenching of fluorescent proteins AmCyan and mOrange2. Mol. Biotechnol. 60, 485–491.
Brakemann, T., Weber, G., Andresen, M., Groenhof, G., Stiel, A.C., Trowitzsch, S., Eggeling, C., Grubmuller, H., Hell, S.W., Wahl, M.C., et al. 2010. Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron. J. Biol. Chem. 285, 14603–14609.
Chen, V. B., Arendall 3rd, W.B., Headd, J.J., Keedy, D.A., Immormino, R.M., Kapral, G.J., Murray, L.W., Richardson, J.S., and Richardson, D.C. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21.
Day, R.N. and Davidson, M.W. 2009. The fluorescent protein palette: tools for cellular imaging. Chem. Soc. Rev. 38, 2887–2921.
Emsley, P. and Cowtan, K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132.
Henderson, J.N., Osborn, M.F., Koon, N., Gepshtein, R., Huppert, D., and Remington, S.J. 2009. Excited state proton transfer in the red fluorescent protein mKeima. J. Am. Chem. Soc. 131, 13212–13213.
Higashino, A., Mizuno, M., and Mizutani, Y. 2016. Chromophore structure of photochromic fluorescent protein Dronpa: Acid-base equilibrium of two Cis configurations. J. Phys. Chem. B 120, 3353–3359.
Jung, G., Wiehler, J., and Zumbusch, A. 2005. The photophysics of green fluorescent protein: influence of the key amino acids at positions 65, 203, and 222. Biophys. J. 88, 1932–1947.
Kim, I.J., Kim, S., Park, J., Eom, I., Kim, S., Kim, J.H., Ha, S.C., Kim, Y.G., Hwang, K.Y., and Nam, K.H. 2016. Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development. FEBS Lett. 590, 2982–2990.
Kogure, T., Karasawa, S., Araki, T., Saito, K., Kinjo, M., and Miyawaki, A. 2006. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence crosscorrelation spectroscopy. Nat. Biotechnol. 24, 577–581.
Kogure, T., Kawano, H., Abe, Y., and Miyawaki, A. 2008. Fluorescence imaging using a fluorescent protein with a large Stokes shift. Methods 45, 223–226.
Krissinel, E. and Henrick, K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797.
Ma, X., Foo, Y.H., and Wohland, T. 2014. Fluorescence cross-correlation spectroscopy (FCCS) in living cells. Methods Mol. Biol. 1076, 557–573.
Nam, K.H., Kwon, O.Y., Sugiyama, K., Lee, W.H., Kim, Y.K., Song, H.K., Kim, E.E., Park, S.Y., Jeon, H., and Hwang, K.Y. 2007. Structural characterization of the photoswitchable fluorescent protein Dronpa-C62S. Biochem. Biophys. Res. Commun. 354, 962–967.
Nienhaus, K., Renzi, F., Vallone, B., Wiedenmann, J., and Nienhaus, G.U. 2006. Chromophore-protein interactions in the anthozoan green fluorescent protein asFP499. Biophys. J. 91, 4210–4220.
Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326.
Park, S.Y., Ha, S.C., and Kim, Y.G. 2017. The protein crystallography beamlines at the Pohang light source II. Biodesign 5, 30–34.
Piatkevich, K.D., Malashkevich, V.N., Almo, S.C., and Verkhusha, V.V. 2010. Engineering ESPT pathways based on structural analysis of LSSmKate red fluorescent proteins with large Stokes shift. J. Am. Chem. Soc. 132, 10762–10770.
Remington, S.J. 2011. Green fluorescent protein: a perspective. Protein Sci. 20, 1509–1519.
Shcherbakova, D.M., Hink, M.A., Joosen, L., Gadella, T.W., and Verkhusha, V.V. 2012. An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J. Am. Chem. Soc. 134, 7913–7923.
Shu, X., Shaner, N.C., Yarbrough, C.A., Tsien, R.Y., and Remington, S.J. 2006. Novel chromophores and buried charges control color in mFruits. Biochemistry 45, 9639–9647.
Tsien, R.Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544.
Vagin, A. and Teplyakov, A. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25.
Violot, S., Carpentier, P., Blanchoin, L., and Bourgeois, D. 2009. Reverse pH-dependence of chromophore protonation explains the large Stokes shift of the red fluorescent protein mKeima. J. Am. Chem. Soc. 131, 10356–10357.
Yang, J., Wang, L., Yang, F., Luo, H., Xu, L., Lu, J., Zeng, S., and Zhang, Z. 2013. mBeRFP, an improved large Stokes shift red fluorescent protein. PLoS One 8, e64849.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at https://doi.org/www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Xu, Y., Hwang, K.Y. & Nam, K.H. Spectral and structural analysis of large Stokes shift fluorescent protein dKeima570. J Microbiol. 56, 822–827 (2018). https://doi.org/10.1007/s12275-018-8319-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-018-8319-5