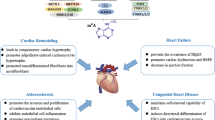
Abstract
Epitranscriptomics is the emerging field of research that comprises the study of epigenetics changes in RNAs. Progressing development in the field of epigenetics has helped to manage and comprehend human diseases. RNA methylation regulates all aspects of RNA functions, which are involved in the pathogenesis of human diseases. Interestingly, RNA m5C methylation is significantly linked to various types of human disease, including cardiovascular diseases (CVD). The m5C methylation is controlled by m5C regulatory proteins, which act as methyltransferase, demethyltransferase, and RNA-binding protein. Dysregulated expression in m5C regulatory proteins is significantly associated with cardiovascular disease, and these regulatory proteins have crucial roles in biological and cellular functions. This review is mainly focused on the role of RNA m5C modification in CVD and mitochondrial dysfunction. Thus, m5C will contribute to discovering the new diagnostic marker and therapeutic target for CVD.
Graphical Abstract




Similar content being viewed by others
Abbreviations
- Asp:
-
Asparagine
- Aza-IP:
-
5-Azacytidine-mediated RNA immunoprecipitation
- CLIP:
-
Cross-linking and immunoprecipitation
- CVD:
-
Cardiovascular disease
- DNMT2:
-
DNA methyltransferase 2
- DS:
-
Dubowitz syndrome
- Gly:
-
Glycine
- Leu:
-
Leucine
- lncRNA:
-
Long noncoding RNA
- m5C:
-
5-Methylcytosine
- m5C-RIP:
-
5-Methylcytosine-RNA immunoprecipitation
- m6A:
-
N6-Methyladenosine
- Me-RIP:
-
Methylated RNA immunoprecipitation
- Met:
-
Methionine
- miRNA:
-
Micro RNA
- mRNA:
-
Messenger RNA
- mt-tRNA:
-
Mitochondrial transfer RNA
- RNMT:
-
RNA methyl transferase
- rRNA:
-
Ribosomal RNA
- SAM:
-
S-Adenosyl-L-methionine
- TAWO-seq:
-
TET-assisted WO-seq
- TCGA:
-
The Cancer Genome Atlas
- TET family:
-
Ten-eleven translocation methylcytosine dioxygenases
- TNF-α:
-
Tumor necrosis factor-α
- TRDMT1:
-
TRNA aspartic acid methyltransferase 1
- tRNA:
-
Transfer RNA
- UTR:
-
Untranslated region
- Val:
-
Valine
- WHO:
-
World Health Organization
References
Anonymous, The top 10 causes of death by WHO, 2020. Retrieved from; https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–54.
Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–9.
Jones P, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41.
Stellos K. The rise of epitranscriptomic era: implications for cardiovascular disease. Cardiovasc Res. 2017;113:e2–3.
Cohn WE. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J Biol Chem. 2017;235:1488–98.
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2018;46:303–7.
Korlach J, Turner SW. Going beyond five bases in DNA sequencing. Curr Opin Struct Biol. 2012;22:251–61.
Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–68.
Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. 2019;10:e1510.
Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–76.
Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, Lusser A. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18(1):1.
Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33.
Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation, and biological functions. Nucleic acids Res. 2010;38:1415–30.
Brown DA, Perry JB, Allen ME, et al. Expert consensus document: mitochondrial function as a therapeutic target in heart failure. Nat Rev Cardiol. 2017;14(4):238–50.
Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–5.
Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). 2019;10(2):102.
Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation, and biological functions. Nucleic acids Res. 2010;38(5):1415–30.
Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. Methylation of tRNA Asp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8.
Reid R, Greene PJ, Santi DV. Exposition of a family of RNA m5C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138–45.
Metodiev MD, Spåhr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10:e1004110.
Van Haute L, Lee SY, McCann BJ, Powell CA, Bansal D, Vasiliauskaitė L, Garone C, Shin S, Kim JS, Frye M, Gleeson JG, Miska EA, Rhee HW, Minczuk M. NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2019;47(16):8720–33.
Sun Z, Xue S, Xu H, Hu X, Chen S, Yang Z, Yang Y, Ouyang J, Cui H. Effects of NSUN2 deficiency on the mRNA 5-methylcytosine modification and gene expression profile in HEK293 cells. Epigenomics. 2019;11:439–53.
Li Y, Li J, Luo M, Zhou C, Shi X, Yang W, Lu Z, Chen Z, Sun N, He J. Novel long noncoding RNA NMR promotes tumor progression via NSUN2 and BPTF in esophageal squamous cell carcinoma. Cancer Lett. 2018;430:57–66.
Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–25.
Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 2006;34:6034–43.
Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–5.
Xue S, Xu H, Sun Z, Shen H, Chen S, Ouyang J, Zhou Q, Hu X, Cui H. Depletion of TRDMT1 affects 5-methylcytosine modification of mRNA and inhibits HEK293 cell proliferation and migration. Biochem Biophys Res Commun. 2019;520:60–6.
Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m5 C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12(4):e1639.
Li X, Meng Y. Expression and prognostic characteristics of m5 C regulators in low-grade glioma. J Cell Mol Med. 2021;25(3):1383–93.
Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–33.
Strobel MC, Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986;6:2663–73.
Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun. 2015;6:6158.
Shen H, Ontiveros RJ, Owens MC, Liu MY, Ghanty U, Kohli RM, Liu KF. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J Biol Chem. 2021;296:100087.
Chistiakov DA, Shkurat TP, Melnichenko AA, Grechko AV, Orekhov AN. The role of mitochondrial dysfunction in cardiovascular disease: a brief review. Ann Med. 2018;50(2):121–7.
Lin H, Miyauchi K, Harada T, Okita R, Takeshita E, Komaki H, Fujioka K, Yagasaki H, Goto YI, Yanaka K, et al. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat Commun. 2018;9:1875.
Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299–329.
Trixl L, Amort T, Wille A, Zinni M, Ebner S, Hechenberger C, Lusser A. RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell Mol Life Sci. 2018;75(8):1483–97.
Spåhr H, Habermann B, Gustafsson CM, Larsson NG, Hallberg BM. Structure of the human MTERF4–NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc Natl Acad Sci USA. 2012;109:15253–8.
Huang T, Chen W, Liu J, Gu N, Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol. 2019;26(5):380–8.
Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, Suzuki T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017;45(12):7401–15.
Zhang B, Jiang H, Dong Z, Sun A, Ge J. The critical roles of m6A modification in metabolic abnormality and cardiovascular diseases. Genes Dis. 2020;8(6):746–58.
Lewinska A, Wnuk M, Grabowska W, Zabek T, Semik E, Sikora E, Bielak-Zmijewska A. Curcumin induces oxidation-dependent cell cycle arrest mediated by SIRT7 inhibition of rDNA transcription in human aortic smooth muscle cells. Toxicol Lett. 2015;233:227–38.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, He C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–99.
Ghanbarian H, Wagner N, Polo B, Baudouy D, Kiani J, Michiels JF, Wagner KD. Dnmt2/Trdmt1 as mediator of RNA polymerase II transcriptional activity in cardiac growth. PLoS ONE. 2016;11(6):e0156953.
Cheng JX, Chen L, Li Y, Cloe A, Yue M, Wei J, Vardiman JW. RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun. 2018;9(1):1163.
Luo Y, Feng J, Xu Q, Wang W, Wang X. NSun2 deficiency protects endothelium from inflammation via mRNA methylation of ICAM-1. Circ Res. 2016;118(6):944–56.
Yuan S, Tang H, Xing J, et al. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol Cell Biol. 2014;34(19):3630–41.
Yin L, Zhu X, Novák P, Zhou L, Gao L, Yang M, Zhao G, Yin K. The epitranscriptome of long noncoding RNAs in metabolic diseases. Clin Chim Acta. 2021;515:80–9.
Martens MA, Wilson SJ, Reutens DC. Williams syndrome: a critical review of the cognitive, behavioral and neuroanatomical phenotype. J Child Psychol Psychiatry. 2008;14:576–608.
Wang N, Tang H, Wang X, Wang W, Feng J. Homocysteine upregulates interleukin-17A expression via NSun2-mediated RNA methylation in T lymphocytes. Biochem Biophys Res Commun. 2017;493(1):94–9. https://doi.org/10.1016/j.bbrc.2017.09.069.
He Y, Zhang H, Yin F, et al. Novel insights into the role of 5-methylcytosine RNA methylation in human abdominal aortic aneurysm. Front Biosci (Landmark Ed). 2021;26(11):1147–65. https://doi.org/10.52586/5016.
Acknowledgements
We acknowledge the contribution of Dr. Anitha Roy, who was involved in the editing and formatting of the manuscript.
Funding
This work was supported by the Science and Engineering Research Board (SERB), Government of India (EEQ/2019/000411).
Author information
Authors and Affiliations
Contributions
K. Balachander undertook literature mining from various reputed databases, drafted the manuscript, and prepared illustrations. Dr. A. Paramasivam and Dr. J. Vijayashree Priyadharsini gave the concept for this article and are responsible for manuscript proofreading and validating the entire manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Associate Editor Junjie Xiao oversaw the review of this article
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Balachander, K., Priyadharsini, J.V., Roy, A. et al. Emerging Role of RNA m5C Modification in Cardiovascular Diseases. J. of Cardiovasc. Trans. Res. 16, 598–605 (2023). https://doi.org/10.1007/s12265-022-10336-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-022-10336-8