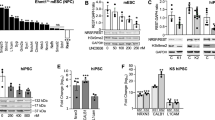
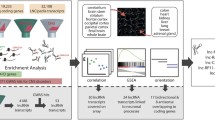
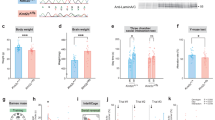
Abstract
Intellectual disability (ID) is a condition characterized by cognitive impairment and difficulties in adaptive functioning. In our research, we identified two de novo mutations (c.955C>T and c.732C>A) at the KDM2A locus in individuals with varying degrees of ID. In addition, by using the Gene4Denovo database, we discovered five additional cases of de novo mutations in KDM2A. The mutations we identified significantly decreased the expression of the KDM2A protein. To investigate the role of KDM2A in neural development, we used both 2D neural stem cell models and 3D cerebral organoids. Our findings demonstrated that the reduced expression of KDM2A impairs the proliferation of neural progenitor cells (NPCs), increases apoptosis, induces premature neuronal differentiation, and affects synapse maturation. Through ChIP-Seq analysis, we found that KDM2A exhibited binding to the transcription start site regions of genes involved in neurogenesis. In addition, the knockdown of KDM2A hindered H3K36me2 binding to the downstream regulatory elements of genes. By integrating ChIP-Seq and RNA-Seq data, we made a significant discovery of the core genes' remarkable enrichment in the MAPK signaling pathway. Importantly, this enrichment was specifically linked to the p38 MAPK pathway. Furthermore, disease enrichment analysis linked the differentially-expressed genes identified from RNA-Seq of NPCs and cerebral organoids to neurodevelopmental disorders such as ID, autism spectrum disorder, and schizophrenia. Overall, our findings suggest that KDM2A plays a crucial role in regulating the H3K36me2 modification of downstream genes, thereby modulating the MAPK signaling pathway and potentially impacting early brain development.







Similar content being viewed by others
Data Availability
RNA-Seq and ChIP-Seq data were deposited in Gene Expression Omnibus (Accession number: GSE229836). All other data associated with this study are shown in the manuscript or Supplementary Materials.
References
Social Security Administration. Change in terminology: “mental retardation” to “intellectual disability” Final rule. Fed Regist 2013, 78: 46499–46502.
Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet 2016, 17: 9–18.
Totsika V, Liew A, Absoud M, Adnams C, Emerson E. Mental health problems in children with intellectual disability. Lancet Child Adolesc Health 2022, 6: 432–444.
Patel DR, Cabral MD, Ho A, Merrick J. A clinical primer on intellectual disability. Transl Pediatr 2020, 9: S23–S35.
van Ool JS, Snoeijen-Schouwenaars FM, Schelhaas HJ, Tan IY, Aldenkamp AP, Hendriksen JGM. A systematic review of neuropsychiatric comorbidities in patients with both epilepsy and intellectual disability. Epilepsy Behav 2016, 60: 130–137.
Stadskleiv K. Cognitive functioning in children with cerebral palsy. Dev Med Child Neurol 2020, 62: 283–289.
Hickman RA, O’Shea SA, Mehler MF, Chung WK. Neurogenetic disorders across the lifespan: from aberrant development to degeneration. Nat Rev Neurol 2022, 18: 117–124.
Maia N, Nabais Sá MJ, Melo-Pires M, de Brouwer APM, Jorge P. Intellectual disability genomics: current state, pitfalls and future challenges. BMC Genom 2021, 22: 909.
Iwase S, Bérubé NG, Zhou Z, Kasri NN, Battaglioli E, Scandaglia M. Epigenetic etiology of intellectual disability. J Neurosci 2017, 37: 10773–10782.
Zahir FR, Brown CJ. Epigenetic impacts on neurodevelopment: pathophysiological mechanisms and genetic modes of action. Pediatr Res 2011, 69: 92R-100R.
Jambhekar A, Dhall A, Shi Y. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol 2019, 20: 625–641.
Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011, 21: 381–395.
Kim JH, Lee JH, Lee IS, Lee SB, Cho KS. Histone lysine methylation and neurodevelopmental disorders. Int J Mol Sci 2017, 18: 1404.
Collins BE, Greer CB, Coleman BC, Sweatt JD. Histone H3 lysine K4 methylation and its role in learning and memory. Epigenetics Chromatin 2019, 12: 7.
Fallah MS, Szarics D, Robson CM, Eubanks JH. Impaired regulation of histone methylation and acetylation underlies specific neurodevelopmental disorders. Front Genet 2021, 11: 613098.
Li D, Xie Y. An evolved human-specific epigenetic mechanism for cortical expansion and gyrification. Neurosci Bull 2021, 37: 1370–1372.
Reichard J, Zimmer-Bensch G. The epigenome in neurodevelopmental disorders. Front Neurosci 2021, 15: 776809.
Scandaglia M, Barco A. Contribution of spurious transcription to intellectual disability disorders. J Med Genet 2019, 56: 491–498.
Larizza L, Finelli P. Developmental disorders with intellectual disability driven by chromatin dysregulation: clinical overlaps and molecular mechanisms. Clin Genet 2019, 95: 231–240.
Marshall P, Bredy TW. Cognitive neuroepigenetics: The next evolution in our understanding of the molecular mechanisms underlying learning and memory? Npj Sci Learn 2016, 1: 16014.
Maity S, Farrell K, Navabpour S, Narayanan SN, Jarome TJ. Epigenetic mechanisms in memory and cognitive decline associated with aging and alzheimer’s disease. Int J Mol Sci. 2021, 22: 12280.
Hamdan FF, Srour M, Capo-Chichi JM, Daoud H, Nassif C, Patry L, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet 2014, 10: e1004772.
de Ligt J, Willemsen MH, van Bon BWM, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012, 367: 1921–1929.
De Vas MG, Boulet F, Joshi SS, Garstang MG, Khan TN, Atla G, et al. Regulatory de novo mutations underlying intellectual disability. Life Sci Alliance 2023, 6: e202201843.
Zhao G, Li K, Li B, Wang Z, Fang Z, Wang X, et al. Gene4Denovo: an integrated database and analytic platform for de novo mutations in humans. Nucleic Acids Res 2020, 48: D913–D926.
Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006, 7: 715–727.
Liu L, Liu J, Lin Q. Histone demethylase KDM2A: Biological functions and clinical values (Review). Exp Ther Med 2021, 22: 723.
Lu B, Wei J, Zhou H, Chen J, Li Y, Ye L, et al. Histone H3K36me2 demethylase KDM2A promotes bladder cancer progression through epigenetically silencing RARRES3. Cell Death Dis 2022, 13: 547.
Chen L, Zhang J, Zou Y, Wang F, Li J, Sun F, et al. Kdm2a deficiency in macrophages enhances thermogenesis to protect mice against HFD-induced obesity by enhancing H3K36me2 at the Pparg locus. Cell Death Differ 2021, 28: 1880–1899.
Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y, et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest 2013, 123: 5231–5246.
Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501: 373–379.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30: 2114–2120.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29: 15–21.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30: 923–930.
Carneiro TN, Krepischi AC, Costa SS, Tojal da Silva I, Vianna-Morgante AM, Valieris R, et al. Utility of trio-based exome sequencing in the elucidation of the genetic basis of isolated syndromic intellectual disability: illustrative cases. Appl Clin Genet 2018, 11: 93–98.
Gao C, Wang X, Mei S, Li D, Duan J, Zhang P, et al. Diagnostic yields of trio-WES accompanied by CNVseq for rare neurodevelopmental disorders. Front Genet 2019, 10: 485.
Gabriel H, Korinth D, Ritthaler M, Schulte B, Battke F, von Kaisenberg C, et al. Trio exome sequencing is highly relevant in prenatal diagnostics. Prenat Diagn 2022, 42: 845–851.
Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 1977, 74: 5463–5467.
Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA, et al. DNA sequencing at 40: past, present and future. Nature 2017, 550: 345–353.
Ball RS. The Gesell developmental schedules: Arnold Gesell (1880–1961). J Abnorm Child Psychol 1977, 5: 233–239.
Weick JP, Held DL, Bonadurer GF 3rd, Doers ME, Liu Y, Maguire C, et al. Deficits in human trisomy 21 iPSCs and neurons. Proc Natl Acad Sci U S A 2013, 110: 9962–9967.
Tsukada YI, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439: 811–816.
Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG Islands recruit a histone H3 lysine 36 demethylase. Mol Cell 2010, 38: 179–190.
Harper JW, Schulman BA. Cullin-RING ubiquitin ligase regulatory circuits: A quarter century beyond the F-box hypothesis. Annu Rev Biochem 2021, 90: 403–429.
Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 2006, 31: 35–40.
Enkhbayar P, Kamiya M, Osaki M, Matsumoto T, Matsushima N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins 2004, 54: 394–403.
Collado-Torres L, Burke EE, Peterson A, Shin J, Straub RE, Rajpurohit A, et al. Regional heterogeneity in gene expression, regulation, and coherence in the frontal cortex and Hippocampus across development and schizophrenia. Neuron 2019, 103: 203-216.e8.
Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 2018, 362: eaat7615.
Gordon A, Yoon SJ, Tran SS, Makinson CD, Park JY, Andersen J, et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat Neurosci 2021, 24: 331–342.
Kim M, Haney JR, Zhang P, Hernandez LM, Wang LK, Perez-Cano L, et al. Brain gene co-expression networks link complement signaling with convergent synaptic pathology in schizophrenia. Nat Neurosci 2021, 24: 799–809.
Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res 2010, 38: 4218–4230.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005, 102: 15545–15550.
Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009, 37: W305–W311.
Leblond CS, Le TL, Malesys S, Cliquet F, Tabet AC, Delorme R, et al. Operative list of genes associated with autism and neurodevelopmental disorders based on database review. Mol Cell Neurosci 2021, 113: 103623.
Finucane BM, Myers SM, Martin CL, Ledbetter DH. Long overdue: Including adults with brain disorders in precision health initiatives. Curr Opin Genet Dev 2020, 65: 47–52.
Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput Struct Biotechnol J 2021, 19: 2960–2967.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15: 550.
Abdi H, Williams LJ. Principal component analysis. Wires. Comput Stat 2010, 2: 433–459.
Rao VS, Srinivas K, Sujini GN, Kumar GN. Protein-protein interaction detection: Methods and analysis. Int J Proteomics 2014, 2014: 147648.
Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 2014, 9: 2329–2340.
Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, et al. Evaluating the clinical validity of gene-disease associations: An evidence-based framework developed by the clinical genome resource. Am J Hum Genet 2017, 100: 895–906.
Li S, Li H, Liu D, Xing Q, Chen X, Zhang H, et al. Identification of novel Mendelian disorders of the epigenetic machinery (MDEMs)-associated functional mutations and neurodevelopmental disorders. QJM 2023, 116: 355–364.
Faundes V, Newman WG, Bernardini L, Canham N, Clayton-Smith J, Dallapiccola B, et al. Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am J Hum Genet 2018, 102: 175–187.
Acknowledgements
We thank Dr. Xiao Mao and Dr. Li Shu for providing the ID case. We also appreciate the technical support from the NHC Key Laboratory of Birth Defects for Research and Prevention at the Hunan Provincial Maternal and Child Health Care Hospital. This work was supported by the National Natural Science Foundation of China (82022024, 31970572, and 31871276), the National Key R&D Project of China (2016YFC1306000 and 2017YFC0908701), the Innovation-driven Project of Central South University (2020CX003), The Natural Science Foundation of Hunan Province (2023JJ40793). NIH grants (U01 MH122591, 1U01MH116489, and 1R01MH110920), and the Postgraduate Scientific Research Innovation Project of Hunan Province (CX20220320).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical Approval
Informed consent was obtained from the adult patient and the guardians of the affected children. This study was approved by the Ethics Committee of the Hunan Provincial Maternal and Child Health Care Hospital, Hunan, China (2020-S003). The authors greatly appreciate the support and cooperation of all patients and their families.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ren, Z., Tang, H., Zhang, W. et al. The Role of KDM2A and H3K36me2 Demethylation in Modulating MAPK Signaling During Neurodevelopment. Neurosci. Bull. (2023). https://doi.org/10.1007/s12264-023-01161-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12264-023-01161-3