Abstract
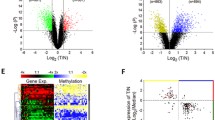
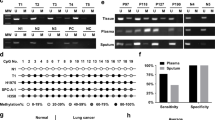
To characterize the DNA methylation as well as exploring the relationship between NID2 methylation and the lung cancer development. Collecting chip data of 9 lung cancer samples and 11 adjacent normal samples from the Gene Expression Omnibus database. Tissues and cells NID2 gene methylation level was measured by methylation-specific PCR. NID2 mRNA level and protein level were validated by Real-Time PCR and Western blot separately. Functional study of lung cancer cells was performed with Cell Counting Kit-8 assay. Colony formation assay, transwell assay, wound healing assay and low cytometry were performed. Finally, NID2 tumorigenesis in vivo was tested in nude mice xenograft models. Microarray analysis outcome present NID2 hypermethylation status in lung cancer tissues. High methylation and low mRNA expression levels of NID2 were detected. After NID2 demethylation or overexpression in cancer cells, cell viability, proliferation, migration as well as invasion ability decreased. Nevertheless, a significant enhancement in apoptosis rate were observed. Overexpressing NID2 or demethylation in lung cancer cells inhibited the tumorigenesis of lung cancer in nude mice. The mRNA and protein level of NID2 in tumors obtained from nude mice xenograft were unanimous with the in vitro assays’ outcome, which significantly decreased after overexpressing NID2 or demethylation. NID2 methylation reduces its expression level and promotes the development of lung cancer.







Similar content being viewed by others
References
Christopoulos P, Kirchner M, Bozorgmehr F, Endris V, Elsayed M, Budczies J, Ristau J, Penzel R, Herth FJ, Heussel CP, Eichhorn M, Muley T, Meister M, Fischer JR, Rieken S, Lasitschka F, Bischoff H, Sotillo R, Schirmacher P, Thomas M, Stenzinger A (2018) Identification of a highly lethal V3(+) TP53(+) subset in ALK(+) lung adenocarcinoma. Int J Cancer. https://doi.org/10.1002/ijc.31893
Morrow JD, Glass K, Cho MH, Hersh CP, Pinto-Plata V, Celli B, Marchetti N, Criner G, Bueno R, Washko G, Choi AMK, Quackenbush J, Silverman EK, DeMeo DL (2018) Human Lung DNA methylation quantitative trait loci Colocalize with chronic obstructive pulmonary disease genome-wide association loci. Am J Respir Crit Care Med 197(10):1275–1284. https://doi.org/10.1164/rccm.201707-1434OC
Chen WQ, Li H, Sun KX, Zheng RS, Zhang SW, Zeng HM, Zou XN, Gu XY, He J (2018) Report of Cancer incidence and mortality in China, 2014. Zhonghua zhong liu za zhi [Chinese journal of oncology] 40(1):5–13. https://doi.org/10.3760/cma.j.issn.0253-3766.2018.01.002
Hong QY, Wu GM, Qian GS, Hu CP, Zhou JY, Chen LA, Li WM, Li SY, Wang K, Wang Q, Zhang XJ, Li J, Gong X, Bai CX, Lung Cancer Group of Chinese Thoracic S, Chinese Alliance Against Lung C (2015) Prevention and management of lung cancer in China. Cancer 121 Suppl 17:3080–3088. https://doi.org/10.1002/cncr.29584
Vlahopoulos S, Adamaki M, Khoury N, Zoumpourlis V, Boldogh I (2018) Roles of DNA repair enzyme OGG1 in innate immunity and its significance for lung cancer. Pharmacol Ther. https://doi.org/10.1016/j.pharmthera.2018.09.004
Feng X, Xie X, Zheng B, Peng C, Zhou H, Qin J (2018) The more potential performance of nidogen 2 methylation by tissue or plasma DNA over brichoalveolar lavage DNA in diagnosis of nonsmall cell lung cancer. J Cancer Res Ther 14(Supplement):S341–S346. https://doi.org/10.4103/0973-1482.235352
Risch A, Plass C (2008) Lung cancer epigenetics and genetics. Int J Cancer 123(1):1–7. https://doi.org/10.1002/ijc.23605
Schubeler D (2015) Function and information content of DNA methylation. Nature 517(7534):321–326. https://doi.org/10.1038/nature14192
Mehrmohamadi M, Mentch LK, Clark AG, Locasale JW (2016) Integrative modelling of tumour DNA methylation quantifies the contribution of metabolism. Nat Commun 7:13666. https://doi.org/10.1038/ncomms13666
Gut P, Verdin E (2013) The nexus of chromatin regulation and intermediary metabolism. Nature 502(7472):489–498. https://doi.org/10.1038/nature12752
Locasale JW (2013) Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13(8):572–583. https://doi.org/10.1038/nrc3557
Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, Nichenametla SN, Locasale JW (2015) Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab 22(5):861–873. https://doi.org/10.1016/j.cmet.2015.08.024
Anderson OS, Sant KE, Dolinoy DC (2012) Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem 23(8):853–859. https://doi.org/10.1016/j.jnutbio.2012.03.003
Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, Van Den Berg D, Laird PW (2011) Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 44(1):40–46. https://doi.org/10.1038/ng.969
Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, Stevenson K, Sougnez C, Wang L, Li S, Kotliar D, Zhang W, Ghandi M, Garraway L, Fernandes SM, Livak KJ, Gabriel S, Gnirke A, Lander ES, Brown JR, Neuberg D, Kharchenko PV, Hacohen N, Getz G, Meissner A, Wu CJ (2014) Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26(6):813–825. https://doi.org/10.1016/j.ccell.2014.10.012
Geng J, Sun J, Lin Q, Gu J, Zhao Y, Zhang H, Feng X, He Y, Wang W, Zhou X, Yu J (2012) Methylation status of NEUROG2 and NID2 improves the diagnosis of stage I NSCLC. Oncol Lett 3(4):901–906. https://doi.org/10.3892/ol.2012.587
Vasiljevic N, Ahmad AS, Beesley C, Thorat MA, Fisher G, Berney DM, Moller H, Yu Y, Lu YJ, Cuzick J, Foster CS, Lorincz AT (2013) Association between DNA methylation of HSPB1 and death in low Gleason score prostate cancer. Prostate Cancer Prostatic Dis 16(1):35–40. https://doi.org/10.1038/pcan.2012.47
Wang Q, Williamson M, Bott S, Brookman-Amissah N, Freeman A, Nariculam J, Hubank MJ, Ahmed A, Masters JR (2007) Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer. Oncogene 26(45):6560–6565. https://doi.org/10.1038/sj.onc.1210472
Chai AW, Cheung AK, Dai W, Ko JM, Ip JC, Chan KW, Kwong DL, Ng WT, Lee AW, Ngan RK, Yau CC, Tung SY, Lee VH, Lam AK, Pillai S, Law S, Lung ML (2016) Metastasis-suppressing NID2, an epigenetically-silenced gene, in the pathogenesis of nasopharyngeal carcinoma and esophageal squamous cell carcinoma. Oncotarget 7(48):78859–78871. https://doi.org/10.18632/oncotarget.12889
Kohfeldt E, Sasaki T, Gohring W, Timpl R (1998) Nidogen-2: a new basement membrane protein with diverse binding properties. J Mol Biol 282(1):99–109. https://doi.org/10.1006/jmbi.1998.2004
Mokkapati S, Bechtel M, Reibetanz M, Miosge N, Nischt R (2012) Absence of the basement membrane component nidogen 2, but not of nidogen 1, results in increased lung metastasis in mice. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society 60(4):280–289. https://doi.org/10.1369/0022155412436586
Ulazzi L, Sabbioni S, Miotto E, Veronese A, Angusti A, Gafa R, Manfredini S, Farinati F, Sasaki T, Lanza G, Negrini M (2007) Nidogen 1 and 2 gene promoters are aberrantly methylated in human gastrointestinal cancer. Mol Cancer 6:17. https://doi.org/10.1186/1476-4598-6-17
Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG (1999) Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res 59(1):67–70
Li B, Lu Q, Song ZG, Yang L, Jin H, Li ZG, Zhao TJ, Bai YF, Zhu J, Chen HZ, Xu ZY (2013) Functional analysis of DNA methylation in lung cancer. Eur Rev Med Pharmacol Sci 17(9):1191–1197
Hsiao SH, Huang TH, Leu YW (2009) Excavating relics of DNA methylation changes during the development of neoplasia. Semin Cancer Biol 19(3):198–208. https://doi.org/10.1016/j.semcancer.2009.02.015
Timpl R, Brown JC (1996) Supramolecular assembly of basement membranes. BioEssays : news and reviews in molecular, cellular and developmental biology 18(2):123–132. https://doi.org/10.1002/bies.950180208
Dong LJ, Hsieh JC, Chung AE (1995) Two distinct cell attachment sites in entactin are revealed by amino acid substitutions and deletion of the RGD sequence in the cysteine-rich epidermal growth factor repeat 2. J Biol Chem 270(26):15838–15843
Wu C, Chung AE, McDonald JA (1995) A novel role for alpha 3 beta 1 integrins in extracellular matrix assembly. J Cell Sci 108(Pt 6):2511–2523
Dedhar S, Jewell K, Rojiani M, Gray V (1992) The receptor for the basement membrane glycoprotein entactin is the integrin alpha 3/beta 1. J Biol Chem 267(26):18908–18914
Author information
Authors and Affiliations
Contributions
Substantial contribution to the conception and design of the work: Jianfeng Wang, Yan Zhao and Hongyan Xu; Acquisition, analysis, and interpretation of the data: Jianfeng Wang, Jun Ma, Feihai Liang and Qingxu Zou; Drafting the manuscript: Jianfeng Wang; Revising the work critically: Fengwu Lin; Final approval of the the version to be published: All authors.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the China-Japan Union Hospital of Jilin University committee. Informed consent was obtained from all individual participants included in the study.
All procedures involving animals were performed in compliance with guidelines of Q China-Japan Union Hospital of Jilin University.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, J., Zhao, Y., Xu, H. et al. Silencing NID2 by DNA Hypermethylation Promotes Lung Cancer. Pathol. Oncol. Res. 26, 801–811 (2020). https://doi.org/10.1007/s12253-019-00609-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-019-00609-0