Abstract
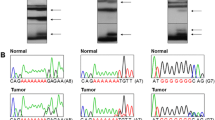
Diminished ANK3 contributes to cell survival by inhibiting detachment-induced apoptosis. TP53BP1 that interacts with p53 and MFN1 that encodes a mitochondrial membrane protein are considered to have tumor suppressor gene (TSG) functions. HACD4 involving fatty acid synthesis and TCPL10 with transcription regulation functions are considered TSGs. Many genes involved in DNA methylations such as LCMT2, RNMT, TRMT6, METTL8 and METTL16 are often perturbed in cancer. The aim of our study was to find whether these genes were mutated in colorectal cancer (CRC). In a genome database, we observed that each of these genes harbored mononucleotide repeats in the coding sequences, which could be mutated in cancers with high microsatellite instability (MSI-H). For this, we studied 124 CRCs for the frameshift mutations of these genes and their intratumoral heterogeneity (ITH). ANK3, HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 harbored 11 (13.9%), 3 (3.8%), 0 (0%), 5 (6.3%), 1 (1.3%), 2 (2.5%), 4 (5.1%), 3 (3.8%), 2 (2.5%) and 2 (2.5%) of 79 CRCs with MSI-H, respectively. However, we found no such mutations in microsatellite stable (MSS) cancers in the nucleotide repeats. There were ITH of the frameshift mutations of ANK3, MFN1 and TP53BP1 in 1 (6.3%), 1 (6.3%) and 1 (6.3%) cases, respectively. Our data exhibit that cancer-related genes ANK3, HACD4, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 harbor mutational ITH as well as the frameshift mutations in CRC with MSI-H. Also, the results suggest that frameshift mutations of these genes might play a role in tumorigenesis through their inactivation in CRC.


Similar content being viewed by others
References
Bennett V, Baines AJ (2011) Spectin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev 81:1353–1392
Hryniewicz-Jankowska A, Czogalla A, Bok E, Sikorsk AF (2002) Ankyrins, multifunctional proteins involved in many cellular pathways. Folia Histochem Cytobiol 40:239–249
Kumar S, Park SH, Cieply B, Schupp J, Killiam E, Zhang F, Rimm DL, Frisch SM (2011) A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition. Mol Cell Biol 31:4036–4051
Glinsky GV, Berezovska O, Glinskii AB (2005) Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 115:1503–1521
Panier S, Boulton SJ (2014) Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15:7–18
Cuella-Martin R, Oliveira C, Lockstone HE, Snellenberg S, Grolmusova N, Chapman JR (2016) 53BP1 integrates DNA repair and p53-dependent cell fate decisions via distinct mechanisms. Mol Cell 64:51–64
Bi J, Huang A, Liu T, Zhang T, Ma H (2015) Expression of DNA damage checkpoint 53BP1 is correlated with prognosis, cell proliferation and apoptosis in colorectal cancer. Int J Clin Exp Pathol 8:6070–6082
Pyakurel A, Savoia C, Hess D, Scorrano L (2015) Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol Cell 58:244–254
Ikeda M, Kanao Y, Yamanaka M, Sakuraba H, Mizutani Y, Igarashi Y, Kihara A (2008) Characterization of four mammalian 3-hydroxyacyl-CoA dehydratases involved in very long-chain fatty acid synthesis. FEBS Lett 582:2435–2440
Zhu S, Wang Z, Zhang Z, Wang J, Li Y, Yao L, Mei Q, Zhang W (2014) PTPLAD2 is a tumor suppressor in esophageal squamous cell carcinogenesis. FEBS Lett 588:981–989
Zuo J, Cai H, Wu Y, Ma H, Jiang W, Liu C, Han D, Ji G, Yu L (2014) TCP10L acts as a tumor suppressor by inhibiting cell proliferation in hepatocellular carcinoma. Biochem Biophys Res Commun 446:61–67
Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, Liang WX, Mi JQ, Song HD, Li KQ, Chen Z, Chen SJ (2011) Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 43:309–315
Imai K, Yamamoto H (2008) Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis 29:673–680
Marusyk A, Almendro V, Polyak K (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 12:323–334
Murphy K, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, Eshleman JR (2006) Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn 8:305–311
Yoo NJ, Kim HR, Kim YR, An CH, Lee SH (2012) Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology 60:943–952
Je EM, Kim MR, Min KO, Yoo NJ, Lee SH (2012) Mutational analysis of MED12 exon 2 in uterine leiomyoma and other common tumors. Int J Cancer 131:E1044–E1047
Jo YS, Choi MR, Song SY, Kim MS, Yoo NJ, Lee SH (2016) Frameshift mutations of HSPA4 and MED13 in gastric and colorectal cancers. Pathol Oncol Res 22:769–772
Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7:131–142
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Nature Reviews Cell 144:646–674
Calin GA, Gafà R, Tibiletti MG, Herlea V, Becheanu G, Cavazzini L, Barbanti-Brodano G, Nenci I, Negrini M, Lanza G (2000) Genetic progression in microsatellite instability high (MSI-H) colon cancers correlates with clinico-pathological parameters: a study of the TGRbetaRII, BAX, hMSH3, hMSH6, IGFIIR and BLM genes. Int J Cancer 89:230–235
Choi YJ, Kim MS, An CH, Yoo NJ, Lee SH (2014) Regional bias of intratumoral genetic heterogeneity of nucleotide repeats in colon cancers with microsatellite instability. Pathol Oncol Res 20:965–971
Choi YJ, Rhee JK, Hur SY, Kim MS, Lee SH, Chung YJ, Kim TM, Lee SH (2017) Intraindividual genomic heterogeneity of high-grade serous carcinoma of the ovary and clinical utility of ascitic cancer cells for mutation profiling. J Pathol 241:57–66
Acknowledgements
This study was supported by grants from Korea Research Foundation (2012R1A5A2047939 and 2017R1A2B2002314).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
None
Rights and permissions
About this article
Cite this article
Yeon, S.Y., Jo, Y.S., Choi, E.J. et al. Frameshift Mutations in Repeat Sequences of ANK3, HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 Genes in Colon Cancers. Pathol. Oncol. Res. 24, 617–622 (2018). https://doi.org/10.1007/s12253-017-0287-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-017-0287-2